Volume 25, Number 2—February 2019
Synopsis
Atypical Cowpox Virus Infection in Smallpox-Vaccinated Patient, France
Cite This Article
Citation for Media
Abstract
We report a case of atypical cowpox virus infection in France in 2016. The patient sought care for thoracic lesions after injury from the sharp end of a metallic guardrail previously stored in the ground. We isolated a cowpox virus from the lesions and sequenced its whole genome. The patient reported that he had been previously vaccinated against smallpox. We describe an alternative route of cowpox virus infection and raise questions about the immunological status of smallpox-vaccinated patients for circulating orthopoxviruses.
The genus Orthopoxvirus (family Poxviridae) is composed of 10 recognized viral species that infect vertebrates and cause serologic cross-reactions. Among the orthopoxviruses, variola virus, which causes smallpox in humans, was associated with the death of millions of persons. An extensive vaccination campaign promoted by the World Health Organization and using multiple vaccinia virus variants (1) during the 1960s and 1970s led to a declaration that smallpox was eradicated in 1980, and vaccination ceased. Most persons born after 1980 have not received smallpox vaccination, and so there is a reduced level of population-based immunity. Coincidentally or not, some zoonotic orthopoxvirus species are re-emerging in an increasing and alarming number of cases worldwide, including vaccinia virus in Brazil and India (2), monkeypox virus in Africa (3,4), cowpox virus in Europe and Asia (5), and novel orthopoxvirus-related strains in the United States (in Alaska and Georgia) (6–8).
Cowpox virus infection in humans causes local cutaneous pustular affections, which may in some cases disseminate and become fatal in immunocompromised patients (9,10). Recent studies showed that cowpox virus is a unique name given to different strains with numerous misnomers (11–13). Rodents seem to be the main reservoirs of cowpox virus (14). Description of cowpox virus infections in cows has been rare in the last years (15). Because cowpox virus can infect a broad range of hosts, viral infections have been reported in cats, monkeys, elephants, llamas, and other vertebrates at zoos in Europe (16,17). Since the 2000s, cowpox virus infections in humans have been frequently associated with direct contact between patients and rodents (18–21), causing lesions mainly on the hands, arms, face, and neck. Human infection can also occur through intermediate hosts, notably by domestic cats, which are commonly infected with cowpox virus through contact with rodents (22). Although infection by fomites is not frequently described for cowpox virus, it is a well-described route of infection for other orthopoxviruses, such as vaccinia virus in Brazil (23).
We report an atypical cowpox virus human infection in France in 2016, in which the patient had a pustular lesion on the laterothoracic area, but reported no direct contact with infected domestic or wild animals. We present our analysis of this novel viral strain, cowpox virus France Amiens 2016, describe its complete genome, review some morphological aspects of its infectious cycle, and discuss the probable way of transmission.
Clinical Examination and Disease Course
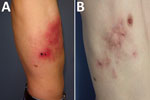
A 45-year-old man, an electrician, had a work accident and was injured by the sharp end of a metallic building site’s guardrail, which was stored in the ground. The lesion was superficial; it affected the derma with little bleeding and did not reach the hypoderma tissue. The laterothoracic wound did not heal and turned into a black eschar with painful cellulitis spreading to the front and upward on the laterothoracic area slowly over 4 weeks (Figure 1, panel A). Multiple treatments were administered by the patient’s general physician with no effect on the course of the disease: amoxicillin (1 g 2×/d), valaciclovir (1 g 3×/d), pristinamycin (1 g 2×/d), ceftriaxone (1 g 4×/d), and doxycycline (100 mg 2×/d).
After 4 weeks, the patient sought further care at Centre Hospitalier Universitaire (CHU) Amiens-Picardie in Amiens, France. He was apyretic, with good general condition and normal vital signs. The whole cellulitis was painful, associated with multiple subcutaneous abscesses and axillary adenopathies. Relevant biologic exams showed increased lymphocyte count (46%, 3.6 G/L); mild hepatitis (aspartate aminotransferase 1.5× the upper limit of normal [ULN], alanine-aminotransferase 1.5× ULN, γ-glutamyl transferase 1.5×ULN); and C-reactive protein (22 mg/L). Electrolytes, prothrombin ratio, partial thromboplastin time, hemoglobin, platelet count, creatinine, procalcitonin, and fibrinogen were normal. Skin biopsy showed a predominantly eosinophilic and neutrophilic necrotizing dermohypodermitis, with intravascular thrombi without vasculitis. Moreover, periodic acid–Schiff (PAS) and Grocott staining showed no pathogens.
At this time, a disease by inoculation was suspected. Results of routine skin biopsy cultures for fungi, bacteria, and mycobacteria were negative, as were intracellular cultures performed on the scar biopsy for Ricketssia spp. Results of molecular detection of herpesviruses, herpes virus 1/2, and varicella zoster virus were negative, as were Bartonella henselae and Franciscela tularensis serologic test results.
The apex of the disease occurred 8 weeks after the initial trauma. Cellulitis grew through the hemithoracic region with purulent discharge from open wounds because of severe delayed healing. The pain required morphine. No wound debridement was needed. Pain spontaneously ceased 4 months after the initial trauma, and the patient was declared healed after 9 months (Figure 1, panel B).
Virus Detection, Isolation, and Production
Similar to the process Ninove et al. described in 2009 (18), a sample was sent to the Institut Hospitalo-Universitaire Méditerranée Infection, Marseille, France, diagnostic laboratory to explore intracellular microorganisms, especially Rickettsia spp. (intracellular bacteria), suspected by the presence of eschar. Nevertheless, we performed other PCR diagnostics at Centre Hospitalier Universitaire Amiens-Picardie. We performed biochemical, hematologic, and serologic examinations using Siemens analyzers (Siemens, https://www.healthcare.siemens.com). We used kits and reagents to detect Bartonella spp., Bartonella henselae, and Bartonella quintana (Eurobio indirect immunofluorescence assay, http://www.eurobio.fr) and the Virion ELISA classic kit (Serion Diagnostics, https://www.serion-diagnostics.de) to detect Francisella tularensis.
At Institut Hospitalo-Universitaire Méditerranée Infection, we performed PCR assays on the cutaneous biopsy taken from the pustular area when the sample was received. To detect orthopoxvirus, we used the primers F-5′-TGATGCAACTCTATCATGTARTCG, R-5′-CAAGACGTCGCTTTTRGCAG, and 6FAM- TGCTTGGTATAAGGAGCCCAATTCCA, targeting the hemagglutinin gene. We conducted real-time PCR for varicella zoster virus and herpesvirus using the ARGENE kit (bioMérieux, https://www.biomerieux-diagnostics.com). We ran PCR for DNA of the 16S RNA gene in parallel, adding to the specific PCR targeting Bartonella spp., Francisella tularensis, and Rickettsia spp. using primers previously reported (24,25).
For culture, we macerated the biopsy sample in Potter-Elvehjem PTFE tissue grinder (Dominique Dutscher Company, shttps://www.dutscher.com) and resuspended it in Hanks’ solution (Thermo Fisher Scientific, http://www.thermofisher.com). Afterward, we inoculated 200 μL of the sample containing 1 mL of Vero (ATCC CCL-81) African green monkey kidney cells at 106 cells/mL onto each of 2 shell vials using 7 mL TRAC bottle (Thermo Fisher Scientific). We placed one at 32°C and the other at 37°C under 5% CO2 atmosphere and observed the vials daily under an inverted microscope to detect any potential cytopathic effect.
For virus production, we prepared 15 flasks of Vero cells in minimum essential medium (MEM) (Thermo Fisher Scientific) with 5% of fetal bovine serum and 1% of glutamine. After the cells reached 80% confluence, we removed the medium and inoculated the monolayer with 5 mL of viral suspension with a multiplicity of infection of 0.01. We incubated the flasks at 37°C for 1 hour for adsorption, then added 20 mL of modified MEM to the flasks and incubated them for 3 days. On the third day, we discarded the supernatant, then washed the cell monolayer 3 times with phosphate buffered saline and removed it using a scraper. After all the flasks were scraped and washed twice to collect the cells, we transferred the contents to 50-mL falcon tubes that were kept on ice.
We then centrifuged the cells at 1,500 rpm for 10 min, discarded the supernatant, and re-suspended the pellet in 10 mL of a sterile lysis buffer (MgCl2 1 mmol/L, Tris 10 mmol/L, pH 7.0 KCl 10 mmol/L). We incubated the suspension for 10 min on ice. We performed mechanical lysis using a sterile tissue grinder (Dominique Dutscher Company, https://www.dutscher.com) (80 cycles on ice). We added 10 mL of 36% sucrose to a plastic centrifugation tube and transferred the viral mixture slowly, avoiding mixing with the sucrose solution (biphasic final solution). We centrifuged the tube at 14,000 rpm for 1 h at 4°C, collected the pellet, and stored it at −80°C in small aliquots.
Micrograph Embedding and Cell Preparation for the Replicative Cycle
Hep2 cells (ATCC accession no. CCL-23) were maintained in culture with MEM modified with 10% of fetal bovine serum. The virus inoculated the Hep2 cell monolayer at a multiplicity of infection of 0.01. We then collected the content after scraping the flask at 32 h postinfection. We followed the same protocol of cell embedding as described by Bou Khalil et al. (26), except that we replaced the Epon resin with LR white resin (Agar Scientific, https://www.agarscientific.com). In brief, we fixed cells for 1 h with 2.5% glutaraldehyde in a 0.1 mmol/L sodium cacodylate buffer and washed them with a mixture of 0.2 mmol/L saccharose and 0.1 mmol/L sodium cacodylate. Postfix was for 1 h with 1% OsO4 diluted in 0.2 mmol/L potassium hexa-cyanoferrate (III) and 0.1 mmol/L sodium cacodylate solution. After washing with distilled water, we gradually dehydrated the cells with ethanol, and then gradually replaced the ethanol with LR white resin. We performed polymerization for 24 h at 60°C. We used a UC7 ultramicrotome (Leica) to obtain ultrathin 70-nm sections and placed them onto HR25 300 mesh copper/rhodium grids (TAAB Laboratories Equipment Ltd., https://www.taab.co.uk). We colored sections with Reynolds solution and obtained electron micrographs on a Tecnai G2 TEM (FEI, https://www.fei.com) operated at 200 keV. We used ImageJ software (https://imagej.nih.gov/ij) to determine particle size.
Genome Sequencing and Assembling
We sequenced genomic cowpox virus DNA (DNAg) on MiSeq technology (Illumina Inc., https://www.illumina.com) with the paired end strategy and barcoded samples to be mixed with 18 other genomic projects prepared with the Nextera XT DNA sample prep kit (Illumina). We quantified the DNAg by high-sensitivity Qubit assay (Life Technologies, https://www.thermofisher.com) to 0.5 ng/µL and performed dilution requiring 1 ng of each genome as input to prepare the paired end library. The tagmentation step fragmented and tagged the DNA. Twelve cycles of limited-cycle PCR amplification completed the tag adapters and introduced dual-index barcodes. After purification on AMPure XP beads (Beckman Coulter Inc., https://www.beckman.com), we normalized the libraries on specific beads in accordance with the Nextera XT protocol (Illumina). We pooled normalized libraries into a single library for sequencing on the MiSeq, then loaded the pooled single-strand library onto the reagent cartridge and then onto the instrument, along with the flow cell. We performed automated cluster generation and paired-end sequencing with dual index reads in a single 39-hour run in 2 × 250-bp.
We obtained total information of 4.3 Gb from a cluster density of 343,000/mm2 with a cluster passing quality control filters of 97.8% (8,331,000 clusters). Within this run, we determined the index representation for cowpox virus to be 9.74%. We filtered the 811,395 paired-end reads according to the read qualities.
We assembled paired-end reads by using CLC genomics workbench version 7.5 (https://www.clcbio.com/) using 64-world size. The genome’s extremities appeared incomplete in comparison to the reference strains. Mapping against cowpox virus France Nancy 2001 (GenBank accession no. HQ420894.1) as reference showed a missing part in the 2 ITRs. We completed the inverted terminal repeat (ITR) regions by PCR followed by sequencing using primers previously designed on primer-Blast (27).
Gene Prediction and Analysis
We computed gene prediction using Genemarks (28) and confirmed by Prodigal (29). We realized a blastp (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) of all predicted proteins against the nonredundant database. To determine average nucleotide value, we compared close phylogenetic strains using the ANI online calculator (https://www.ezbiocloud.net/tools/ani) based on the OrthoANI algorithm (30). Proteinortho (31) was used to determine best reciprocal hits using coverage of 80%, identity 20%, and an E-value cutoff established at 0.01. The genome sequenced in this study is available on the EMBLD/EBI website (accession no. LT883663).
Phylogenetic Analysis
We computed alignments using MAFFT version 7 (32) with fast Fourier transform, a heuristic progressive method (FFT-NS2), on a 73-nt complete genome obtained from the Virus Pathogen Database and Analysis Resource (https://www.viprbrc.org/brc/home.spg). Alignments were manually controlled on MEGA version 6.0 (33).We used the FastTree program (34) to construct a maximum-likelihood tree using standard parameters with the Jukes-Cantors method for the nucleotide distances calculation with 1,000 local resamples (Shimodaira-Hasegawa test). We visualized trees by using iTol (35).
Isolation of the Cowpox Virus and Clinical Context
All bacterial PCR were negative and excluded any DNA bacterial presence. However, specific orthopoxvirus PCR was positive. In parallel, the inoculation on Vero cell showed a typical cytopathic effect after 4 days at 32°C and 37°C. Because cowpox is a notifiable disease, we reported the case to government authorities.
All vaccinations for the patient were up to date; he had received an injection against smallpox with vaccinia virus strain Lister when he was 1 year of age. Following governmental recommendations, no booster vaccination was given after the first injection. The patient did not report other chronic diseases, allergies, or addictions. He reported having a domestic cat at home who also lived outside. The patient’s cat was examined by a veterinarian and showed no sign of cowpox infection during this period. The patient is sure he was not scratched by his cat before the work accident occurred.
Cowpox Virus Strain Genomic Analysis
We obtained a 219,385-bp genome with a GC content estimated at 33.6%. The gene prediction established the number of open reading frames at 214. Altogether, 212/214 predicted proteins had results in the nonredundant database; most (191) best-hit results were obtained for cowpox virus from various previously described strains, 9 for vaccinia virus, 4 for variola virus, 3 for monkeypoxvirus, 2 for ectromelia virus, and 1 each for horsepox, camelpox, and taterapox. The 2 other genes were considered as ORFan (Open Reading Frames without detectable homologues in other lineages), located in the ITR regions. We decided to explore the phylogeny of this new isolate.
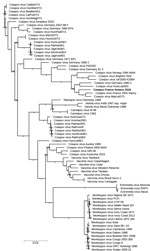
A central part of the orthopoxvirus genome is extremely conserved. Regarding the recent proposed classifying elements of cowpox virus (12,13,36,37), we performed phylogenetic analysis on the available whole genome. We observed a subtype containing the novel strain, cowpox virus France Amiens 2016, along with the Nancy 2001 strain, the MarLei07/1, the HumLue09/1, and the Germany 1990 strains (Figure 2). Using the OrthoANI algorithm, we observed that France Amiens 2016 presented the highest similarity, 98.54%, with the cowpox France Nancy 2001 virus (Appendix Table 1). Moreover, the amino acid comparison of the main functional proteins showed a clear difference between the reference cowpox virus Brighton red strain and the other reported strains of the same cluster (Appendix Table 2). Taking all of these elements into consideration, we believe that cowpox virus France Amiens 2016 represents a new original strain clustering with the proposed E3 subclade in Europe (12).
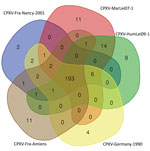
Among the orthopoxvirus genus, the genomes’ evolution seems to be driven by numerous deletions affecting the number of predicted proteins, which could lead to a reduction of the genome length (38). To investigate predicted proteins in this group, we defined the cluster of orthologous proteins by reciprocal best hit. Five genomes of the defined E3 clade shared 193 predicted proteins (Figure 3). For the other clusters, we detected only duplicate proteins and variations in the size of the proteins by modifications of the start codon or by the modification of the stop codon (data not shown).
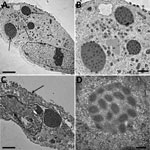
To complete the description of this new isolate, we explored the morphological features in the viral replicative stage. Electron microscopy showed a typical A type inclusion (Figure 4) in the cytoplasm, classifying the cowpox France Amiens 2016 virus in the V+ subtype (39).
The story of orthopoxviruses seems to be clear, but many clues are still missing and ambiguous, where the literature shows much divergent data regarding traced sources, reservoirs, and contamination routes and tools. We are aware that rats, mice, raccoons, and field and bank voles are all recognized as susceptible to cowpox virus, and some of these animals can be associated with human cowpox virus infections. Moreover, serologic evidence highlights wide cowpox virus distribution in rodents and in cats (40,41). Bovids were considered a reservoir before the studies of Baxby (14,42) hypothesized that infections in cows and humans occurred when contaminated brambles and barbed wire were in the proximity of the cattle, but no data confirmed this last assertion. Nevertheless, it is important to highlight that transmission of other orthopoxviruses by fomites is well documented, especially for vaccinia virus, which can be transmitted among cattle by milking devices, as in Brazil (43).
This transmission by fomites should be integrated into upcoming studies with the availability of improved tools, notably in molecular biology, cell culture, and genome sequencing. Nevertheless, the case we report highlights that cowpox virus infection can be misdiagnosed by an atypical clinical presentation, resembling varicella zoster lesions or even noninfectious related rashes. In addition, for this case, it was not possible to establish an epidemiologic link between the patient and the typical sources of infection. Because the tool that caused the wound was kept on the ground, it is possible that it had been contaminated by contact with rodent or cat urine or feces. As is the case in almost all reported infection cases where the diagnosis occurs months later, it becomes difficult to retrace and investigate the route, initial host, or reservoir at an early time of infection. Another scenario is contact between patient and cat after the patient injury, something that was not reported by the patient, but this second potential route of infection appeared doubtful when we examined the co-localization between the injury with the guardrail and the eschar.
Smallpox vaccination is known to confer cross-immunity against other orthopoxviruses (44,45) with a high rate of success when the injection was done in preexposure conditions compared with postexposition. However, despite the patient’s smallpox vaccination, novel infections by orthopoxvirus (46) could have occurred. This cowpox infection is the result of a nonprotective status for the patient; possible causes include an absence of cross-reaction between vaccinia strain and cowpox virus subclade E3 or too long a period between immunization and exposition (nearly 44 years). We have no access to serologic tests or other analyses that were performed before the infection that could be used to confirm one of these hypotheses over another.
Finally, this case is the third reported in Europe within a year (10,47). As confirmed by genomic comparison in some geographic clusters, various strains seem to be circulating in wildlife in Europe, which is alarming because the diagnosis is always delayed when orthopoxvirus infection is not an initial suspect. In this context, the emergence and reemergence of diverse strains of orthopoxvirus must be seriously taken into consideration (48), as should the lack of investigation of potential outbreaks. Multiplying new genome sequences associated with exhaustive clinical reports seems to be an appropriate strategy (12,49) to explore cowpox virus diversity and variants in Europe in general and France in particular.
Dr. Andreani is a PharmD-PhD student in the INSERM program at Aix-Marseille University, Marseille, France. His research interests include virology, new isolates, and genomic characterization. Dr. Arnault is a physician at University Hospital, Amiens, France. His major clinical interest is the management of skin cancer including sonographic early diagnosis, surgery, and systemic treatment.
Acknowledgments
We thank Claire Andréani for her help with the English correction.
This work was supported by the Government of France under the Investissements d’avenir (Investments for the Future) program managed by the Agence Nationale de la Recherche (ANR; National Agency for Research) (reference: Méditerranée Infection 10-IAHU-03).
References
- Qin L, Favis N, Famulski J, Evans DH. Evolution of and evolutionary relationships between extant vaccinia virus strains. J Virol. 2015;89:1809–24. DOIPubMedGoogle Scholar
- Abrahão JS, Campos RK, Trindade GS, Guimarães da Fonseca F, Ferreira PCP, Kroon EG. Outbreak of severe zoonotic vaccinia virus infection, Southeastern Brazil. Emerg Infect Dis. 2015;21:695–8. DOIPubMedGoogle Scholar
- Rimoin AW, Mulembakani PM, Johnston SC, Lloyd Smith JO, Kisalu NK, Kinkela TL, et al. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc Natl Acad Sci U S A. 2010;107:16262–7. DOIPubMedGoogle Scholar
- Reynolds MG, Carroll DS, Karem KL. Factors affecting the likelihood of monkeypox’s emergence and spread in the post-smallpox era. Curr Opin Virol. 2012;2:335–43. DOIPubMedGoogle Scholar
- Vorou RM, Papavassiliou VG, Pierroutsakos IN. Cowpox virus infection: an emerging health threat. Curr Opin Infect Dis. 2008;21:153–6. DOIPubMedGoogle Scholar
- Vora NM, Li Y, Geleishvili M, Emerson GL, Khmaladze E, Maghlakelidze G, et al. Human infection with a zoonotic orthopoxvirus in the country of Georgia. N Engl J Med. 2015;372:1223–30. DOIPubMedGoogle Scholar
- Springer YP, Hsu CH, Werle ZR, Olson LE, Cooper MP, Castrodale LJ, et al. Novel orthopoxvirus infection in an Alaska resident. Clin Infect Dis. 2017;64:1737–41. DOIPubMedGoogle Scholar
- Gao J, Gigante C, Khmaladze E, Liu P, Tang S, Wilkins K, et al. Genome sequences of Akhmeta virus, an early divergent Old World orthopoxvirus. Viruses. 2018;10:252. DOIPubMedGoogle Scholar
- Eis-Hübinger AM, Gerritzen A, Schneweis KE, Pfeiff B, Pullmann H, Mayr A, et al. Fatal cowpox-like virus infection transmitted by cat. Lancet. 1990;336:880. DOIPubMedGoogle Scholar
- Gazzani P, Gach JE, Colmenero I, Martin J, Morton H, Brown K, et al. Fatal disseminated cowpox virus infection in an adolescent renal transplant recipient. Pediatr Nephrol. 2017;32:533–6. DOIPubMedGoogle Scholar
- Carroll DS, Emerson GL, Li Y, Sammons S, Olson V, Frace M, et al. Chasing Jenner’s vaccine: revisiting cowpox virus classification. PLoS One. 2011;6:e23086. DOIPubMedGoogle Scholar
- Mauldin MR, Antwerpen M, Emerson GL, Li Y, Zoeller G, Carroll DS, et al. Cowpox virus: What’s in a Name? Viruses. 2017;9:101. DOIPubMedGoogle Scholar
- Franke A, Pfaff F, Jenckel M, Hoffmann B, Höper D, Antwerpen M, et al. Classification of cowpox viruses into several distinct clades and identification of a novel lineage. Viruses. 2017;9:142. DOIPubMedGoogle Scholar
- Baxby D. Is cowpox misnamed? A review of 10 human cases. BMJ. 1977;1:1379–81. DOIPubMedGoogle Scholar
- Meyer H, Schay C, Mahnel H, Pfeffer M. Characterization of orthopoxviruses isolated from man and animals in Germany. Arch Virol. 1999;144:491–501. DOIPubMedGoogle Scholar
- Baxby D, Shackleton WB, Wheeler J, Turner A. Comparison of cowpox-like viruses isolated from European zoos. Brief report. Arch Virol. 1979;61:337–40. DOIPubMedGoogle Scholar
- Baxby D, Ashton DG, Jones DM, Thomsett LR. An outbreak of cowpox in captive cheetahs: virological and epidemiological studies. J Hyg (Lond). 1982;89:365–72. DOIPubMedGoogle Scholar
- Ninove L, Domart Y, Vervel C, Voinot C, Salez N, Raoult D, et al. Cowpox virus transmission from pet rats to humans, France. Emerg Infect Dis. 2009;15:781–4. DOIPubMedGoogle Scholar
- Campe H, Zimmermann P, Glos K, Bayer M, Bergemann H, Dreweck C, et al. Cowpox virus transmission from pet rats to humans, Germany. Emerg Infect Dis. 2009;15:777–80. DOIPubMedGoogle Scholar
- Vogel S, Sárdy M, Glos K, Korting HC, Ruzicka T, Wollenberg A. The Munich outbreak of cutaneous cowpox infection: transmission by infected pet rats. Acta Derm Venereol. 2012;92:126–31. DOIPubMedGoogle Scholar
- Ducournau C, Ferrier-Rembert A, Ferraris O, Joffre A, Favier A-L, Flusin O, et al. Concomitant human infections with 2 cowpox virus strains in related cases, France, 2011. Emerg Infect Dis. 2013;19:1996–9. DOIPubMedGoogle Scholar
- Żaba R, Jałowska M, Kowalczyk MJ, Bowszyc-Dmochowska M, Adamski Z, Szkaradkiewicz A. Cowpox virus infection in a child after contact with a domestic cat: a case report. New Microbiol. 2017;40:148–50.PubMedGoogle Scholar
- Wood JP, Choi YW, Wendling MQ, Rogers JV, Chappie DJ. Environmental persistence of vaccinia virus on materials. Lett Appl Microbiol. 2013;57:399–404. DOIPubMedGoogle Scholar
- Angelakis E, Roux V, Raoult D, Rolain JM. Real-time PCR strategy and detection of bacterial agents of lymphadenitis. Eur J Clin Microbiol Infect Dis. 2009;28:1363–8. DOIPubMedGoogle Scholar
- Sokhna C, Mediannikov O, Fenollar F, Bassene H, Diatta G, Tall A, et al. Point-of-care laboratory of pathogen diagnosis in rural Senegal. PLoS Negl Trop Dis. 2013;7:e1999. DOIPubMedGoogle Scholar
- Bou Khalil JY, Benamar S, Di Pinto F, Blanc-Tailleur C, Raoult D, La Scola B. Protochlamydia phocaeensis sp. nov., a new Chlamydiales species with host dependent replication cycle. Microbes Infect. 2017;19:343–50. DOIPubMedGoogle Scholar
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. DOIPubMedGoogle Scholar
- Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–18. DOIPubMedGoogle Scholar
- Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. DOIPubMedGoogle Scholar
- Lee I, Ouk Kim Y, Park S-C, Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–3. DOIPubMedGoogle Scholar
- Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. DOIPubMedGoogle Scholar
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. DOIPubMedGoogle Scholar
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50. DOIPubMedGoogle Scholar
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242-5. DOIPubMedGoogle Scholar
- Dabrowski PW, Radonić A, Kurth A, Nitsche A. Genome-wide comparison of cowpox viruses reveals a new clade related to Variola virus. PLoS One. 2013;8:e79953. DOIPubMedGoogle Scholar
- Babkin IV, Babkina IN. The origin of the variola virus. Viruses. 2015;7:1100–12. DOIPubMedGoogle Scholar
- Hatcher EL, Hendrickson RC, Lefkowitz EJ. Identification of nucleotide-level changes impacting gene content and genome evolution in orthopoxviruses. J Virol. 2014;88:13651–68. DOIPubMedGoogle Scholar
- Hoffmann D, Franke A, Jenckel M, Tamošiūnaitė A, Schluckebier J, Granzow H, et al. Out of the reservoir: phenotypic and genotypic characterization of a novel cowpox virus isolated from a common vole. J Virol. 2015;89:10959–69. DOIPubMedGoogle Scholar
- Nowotny N. [Serologic studies of domestic cats for potential human pathogenic virus infections from wild rodents] [in German]. Zentralbl Hyg Umweltmed. 1996;198:452–61.PubMedGoogle Scholar
- Tryland M, Sandvik T, Holtet L, Nilsen H, Olsvik O, Traavik T. Antibodies to orthopoxvirus in domestic cats in Norway. Vet Rec. 1998;143:105–9. DOIPubMedGoogle Scholar
- Kroon EG, Mota BEF, Abrahão JS, da Fonseca FG, de Souza Trindade G. Zoonotic Brazilian Vaccinia virus: from field to therapy. Antiviral Res. 2011;92:150–63. DOIPubMedGoogle Scholar
- Keckler MS, Reynolds MG, Damon IK, Karem KL. The effects of post-exposure smallpox vaccination on clinical disease presentation: addressing the data gaps between historical epidemiology and modern surrogate model data. Vaccine. 2013;31:5192–201. DOIPubMedGoogle Scholar
- Sánchez-Sampedro L, Perdiguero B, Mejías-Pérez E, García-Arriaza J, Di Pilato M, Esteban M. The evolution of poxvirus vaccines. Viruses. 2015;7:1726–803. DOIPubMedGoogle Scholar
- Eder I, Vollmar P, Pfeffer M, Naether P, Rodloff AC, Meyer H. Two distinct clinical courses of human cowpox, Germany, 2015. Viruses. 2017;9:375. DOIPubMedGoogle Scholar
- Grönemeyer L-L, Baltzer A, Broekaert S, Schrick L, Möller L, Nitsche A, et al. Generalised cowpox virus infection. Lancet. 2017;390:1769. DOIPubMedGoogle Scholar
- Shchelkunov SN. An increasing danger of zoonotic orthopoxvirus infections. PLoS Pathog. 2013;9:e1003756. DOIPubMedGoogle Scholar
- Duraffour S, Mertens B, Meyer H, van den Oord JJ, Mitera T, Matthys P, et al. Emergence of cowpox: study of the virulence of clinical strains and evaluation of antivirals. PLoS One. 2013;8:e55808. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: January 07, 2019
1These first authors contributed equally to this article.
Table of Contents – Volume 25, Number 2—February 2019
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Didier Raoult or Bernard La Scola, Aix-Marseille Université, IHU Méditerranée Infection, 19-21 Bd Jean Moulin 13005 Marseille, France;
Top