Volume 25, Number 3—March 2019
Research
SNP-IT Tool for Identifying Subspecies and Associated Lineages of Mycobacterium tuberculosis Complex
Figure 2
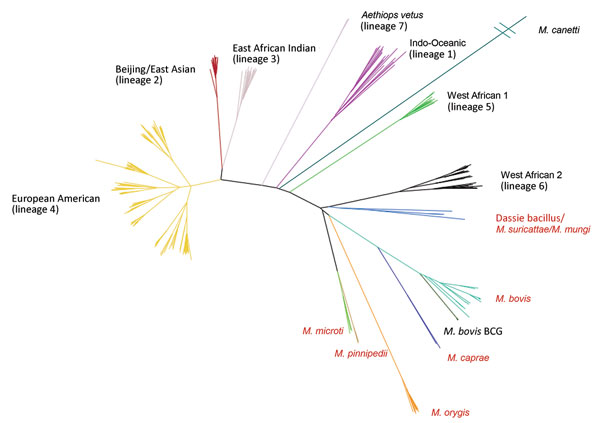
Figure 2. Maximum-likelihood tree built from 70,144 informative positions from whole-genome sequences of all 323 Mycobacterium tuberculosis complex samples in the calibration set for the new SNPs to Identify TB tool. Lineages are of Mycobacterium tuberculosis. Red text denotes animal subspecies. BCG, bacillus Calmette–Guérin; SNP, single-nucleotide polymorphism.
1These authors contributed equally to this article.
2Joint senior authors.
Page created: February 19, 2019
Page updated: February 19, 2019
Page reviewed: February 19, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.