Simplified Model to Survey Tuberculosis Transmission in Countries Without Systematic Molecular Epidemiology Programs
Juan Domínguez
1, Fermín Acosta
1, Laura Pérez-Lago, Dilcia Sambrano, Victoria Batista, Carolina De La Guardia, Estefanía Abascal, Álvaro Chiner-Oms, Iñaki Comas, Prudencio González, Jaime Bravo, Pedro Del Cid, Samantha Rosas, Patricia Muñoz, Amador Goodridge
2
, and Darío García de Viedma
2
Author affiliations: Instituto de Investigaciones Científicas y Servicios de Alta Tecnología, City of Knowledge, Panama (J. Domínguez, F. Acosta, D. Sambrano, V. Batista, C. De La Guardia, A. Goodridge); Instituto Conmemorativo Gorgas de Estudios de la Salud, Panama City, Panama (J. Domínguez, P. González, J. Bravo, P. Del Cid, S. Rosas); Hospital General Universitario Gregorio Marañón, Madrid, Spain (F. Acosta, L. Pérez-Lago, E. Abascal, P. Muñoz, D. García de Viedma); Instituto de Investigación Sanitaria Gregorio Marañón, Madrid, Spain (F. Acosta, L. Pérez-Lago, E. Abascal, P. Muñoz, D. García de Viedma); Centro Superior de investigación en Salud Pública (FISABIO)–Universitat de València, Valencia, Spain (Á. Chiner-Oms); Instituto de Biomedicina de Valencia Consejo Superior de Investigaciones Científicas, Valencia (I. Comas); Centro de Investigación Biomédica en Red en Epidemiología y Salud Pública, Madrid (I. Comas); Universidad Complutense de Madrid, Madrid (P. Muñoz); Centro de Investigación Biomédica en Red Enfermedades Respiratorias, Madrid (P. Muñoz, D. García de Viedma)
Main Article
Figure 2
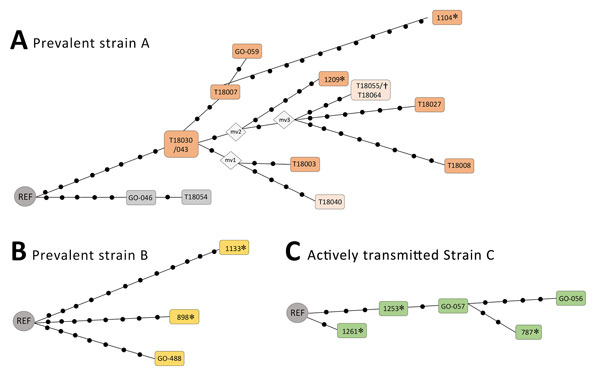
Figure 2. Networks of relationships based on whole-genome sequencing data of prevalent clusters (A, B) and active transmission cluster (C) in testing of a simplified model to survey tuberculosis transmission using data from patients and controls in Panama and Colon provinces, Panama, 2015. Each dot corresponds to a single-nucleotide polymorphism (SNP). In (A), when 2 isolates are included in the same box, they showed no SNPs between them; the isolates within boxes with different colors show mycobacterial interspersed repetitive units–variable number of tandem repeats patterns with single-locus variations between them. Mv, median vector corresponding to nonsampled nodes; REF, reference; *Isolates from 2015 used to design the allele-specific oligonucleotide–PCRs; †strain identified in an additional analysis in Madrid (out of the prospective study applying the strain specific PCRs in Panamá).
Main Article
Page created: February 19, 2019
Page updated: February 19, 2019
Page reviewed: February 19, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
