Volume 25, Number 3—March 2019
Research
Multicenter Study of Cronobacter sakazakii Infections in Humans, Europe, 2017
Figure 2
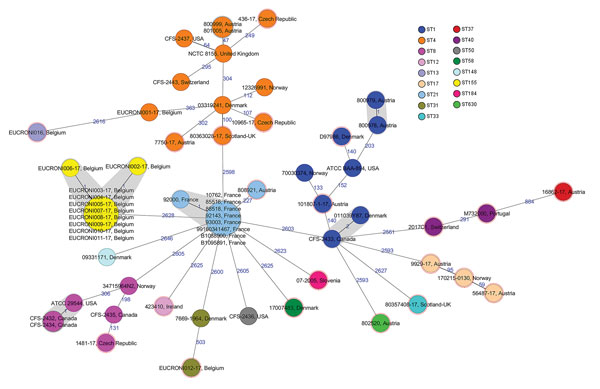
Figure 2. Minimum-spanning tree of 59 Cronobacter sakazakii isolates, including 21 human isolates from 2017 and 38 historical human isolates, from 11 countries in Europe. Each circle represents isolates with an allelic profile based on the core genome multilocus sequence type, which consists of 2,831 alleles. Blue numbers indicate the allelic differences between isolates; isolates with closely related genotypes are shaded in gray. Isolates were colored according to classical multilocus sequence type, labeled with the country of isolation and the respective sample identification. Nodes encircled with a dotted red line were collected in 2017. Ireland additionally provided 7 historical isolates originating from Canada (n = 4), United States (n = 2), and Switzerland (n = 1). For comparison, sequence data of reference strains ATCC BAA-894 (United States, ST1), ATCC29544 (United States, ST8), NCTC 8155 (United Kingdom, ST4) were included. ST, sequence type.
1Members of the EUCRONI study group are listed at the end of this article.