Volume 25, Number 3—March 2019
Research
Multicenter Study of Cronobacter sakazakii Infections in Humans, Europe, 2017
Cite This Article
Citation for Media
Abstract
Cronobacter sakazakii has been documented as a cause of life-threating infections, predominantly in neonates. We conducted a multicenter study to assess the occurrence of C. sakazakii across Europe and the extent of clonality for outbreak detection. National coordinators representing 24 countries in Europe were requested to submit all human C. sakazakii isolates collected during 2017 to a study center in Austria. Testing at the center included species identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, subtyping by whole-genome sequencing (WGS), and determination of antimicrobial resistance. Eleven countries sent 77 isolates, including 36 isolates from 2017 and 41 historical isolates. Fifty-nine isolates were confirmed as C. sakazakii by WGS, highlighting the challenge of correctly identifying Cronobacter spp. WGS-based typing revealed high strain diversity, indicating absence of multinational outbreaks in 2017, but identified 4 previously unpublished historical outbreaks. WGS is the recommended method for accurate identification, typing, and detection of this pathogen.
Cronobacter sakazakii is a motile, gram-negative, rod-shaped opportunistic pathogen of the family Enterbacteriaceae (1). In 2007, organisms previously classified as Enterobacter sakazakii were reassigned to the new genus Cronobacter, which now consists of 7 species: C. sakazakii, C. condimenti, C. dublinensis, C. malonaticus, C. muytjensii, C. turicensis, and C. universalis (2,3). C. sakazakii has been isolated from various environments (e.g., domestic environments and manufacturing plants), clinical sources (e.g., cerebrospinal fluid, blood, and sputum), food (e.g., cheese, meat, and vegetables), and animals (e.g., rats and flies) (4,5).
Most reported cases of illness caused by C. sakazakii are in infants <2 months old (6,7). Premature infants and infants with underlying medical conditions are at the greatest risk for illness. Numerous outbreaks caused by C. sakazakii have been traced to contaminated powdered infant formula (8). Powdered infant formula is not a sterile product, and the ability of C. sakazakii to tolerate dry conditions enables it to survive for long periods in the final powdered product (9).
The screening of food (particularly powdered formula) was proposed to reduce the risk to neonatal and infant health (10,11). The most common syndromes of foodborne infection in infants include necrotizing enterocolitis (NEC), bacteremia, and meningitis (12,13). Examples of outbreaks of illness in hospital neonatal units caused by C. sakazakii associated with powdered infant formula have been compiled by Iversen and Forsythe (6) and by Lund (8).
A few cases of illness (usually nongastrointestinal) in adults caused by C. sakazakii have been reported. In most of these cases the adults had underlying diseases, and no evidence of foodborne transmission was reported (14,15).
We performed a multicenter study of C. sakazakii infections in humans (EUCRONI) to determine the occurrence of C. sakazakii in clinical microbiology laboratories across Europe. We also assessed the extent of clonality for human C. sakazakii isolates.
Study Design
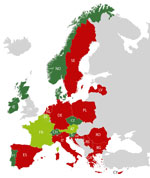
EUCRONI consisted of national coordinators (EUCRONI study group members) from 24 countries in Europe. Coordinators had to actively approach all medical microbiology laboratories to collect human C. sakazakii isolates (1 per patient) in their respective countries during 2017. Human historical isolates (with isolation dates before 2017) were also accepted. The 24 participating countries were arbitrarily chosen to reflect a wide geographic and socioeconomic range (Figure 1). Isolates were transferred to the study center (Austrian Agency for Health and Food Safety, Vienna, Austria) for whole-genome sequencing (WGS), matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) analysis, and antimicrobial drug susceptibility testing. We submitted data capture forms to national coordinators to collect the following demographic data: patient age and sex, patient status (colonized or infected), specimen source, type of healthcare facility requesting the microbiologic culture, and date of specimen collection.
Species Identification and DNA Extraction
We cultured isolates on Columbia blood agar plates (bioMérieux, https://www.biomerieux.com/) overnight at 37°C. We performed species identification by using MALDI-TOF Biotyper (Bruker, https://www.bruker.com) and MBT Compass IVD 4.1.60 (Bruker) according to the manufacturer’s instructions. We conducted isolation, quantification, and WGS of genomic DNA according to methods described by Lepuschitz et al. (16). We used Sequencing Coverage Calculator (https://www.illumina.com) for calculation of a desired mean coverage of >80-fold.
WGS Data Analysis
We de novo assembled raw reads by using SPAdes version 3.9.0 (17) and processed them in SeqSphere+ (Ridom GmbH, https://www.ridom.de) for bacterial typing. We deposited the genome sequences in the PubMLST Cronobacter database (https://pubmlst.org/Cronobacter) under accession nos. 2403 and 2495–2552. To determine the core genome multilocus sequence type (cgMLST) gene set, we performed a genome-wide gene-by-gene comparison by using the MLST+ target definer function of SeqSphere+ as described previously (18) with default parameters and the complete genome of C. sakazakii strain ATCC BAA-894 (19) as reference genome, all complete C. sakazakii genomes available at GenBank, 8 isolates retrieved from whole-genome shotgun sequencing projects, and 4 C. sakazakii isolates sequenced at the Austria study center as query genomes. We extracted sequences of the 7 genes comprising the allelic profile of the classical MLST scheme and queried them against the C. sakazakii MLST database (1), assigning classical sequence types (STs) in silico. We obtained additional species confirmation by using JSpeciesWS (20) and ribosomal MLST (21). We included 23 C. sakazakii historical isolates from 4 different outbreaks (F. Allerberger, 2016; F. Barbut, 2010–2016; G. Feierl, 2009; D. Piérard, 1997–1998, all unpub. data; Appendix Table 1 and 3 reference strains, ATCC BAA-894 (19), ATCC29544 (PRJNA224116), and NCTC 8155 (PRJNA224116), to determine the level of microevolution.
Antimicrobial Resistance Testing
We performed in vitro susceptibility testing with the VITEK 2 Compact System (bioMérieux) and interpreted the VITEK 2 AST196 card according to European Committee on Antimicrobial Susceptibility Testing criteria for Enterobacteriaceae (Clinical Breakpoint Tables version 8.0, http://www.eucast.org/ast_of_bacteria/previous_versions_of_documents). For detection of antibiotic resistance genes, we used the Comprehensive Antibiotic Resistance Database (22) with default settings “perfect” and “strict” for sequence analysis. We tested isolates in SeqSphere+ for Cronobacter-specific variant ampC (e.g., CSA-1, CSA-2, CMA-1, and CMA-2) (23).
Strain Collection and Primary Species Identification
During the study period, 11 of 24 national coordinators (Figure 1) provided 77 presumptive C. sakazakii isolates previously identified by conventional biochemical testing, local MALDI-TOF MS analysis (Bruker Biotyper and VITEK MS), locally performed Cronobacter genus- and species-specific PCRs, or 16S rRNA gene sequence analysis. These 77 isolates consisted of 36 human isolates from 2017 and 41 historical human isolates obtained during 1964–2016. The participating laboratories, using local conventional phenotypical methods or local MALDI-TOF MS analysis, incorrectly identified 18 (23.4%) of 77 human isolates as C. sakazakii.
MALDI-TOF MS analysis in the study center identified 69 of 77 isolates as C. sakazakii; 1 isolate from 2017 yielded low-confidence identification (log[score] value 1.70–1.99). We assigned 7 clinical isolates from 2017 and 1 historical clinical isolate from 2005 to other species (Table 1). The WGS-based species identification using JSpeciesWS and rMLST confirmed MALDI-TOF MS identification results in all but 10 of the 69 isolates. WGS indicated that 5 isolates were C. malonaticus, 2 were C. turicensis, 1 was C. dublinensis, 1 was C. universalis, and 1 was Siccibacter turicensis (Table 1; Appendix Table 1).
Human C. sakazakii Isolates Collected in 2017
In total, 21 C. sakazakii isolates from 21 patients were collected in 2017 in 9 participating countries in Europe. Case-fatality ratio (within 30 days after specimen collection) was 2 of 21 case-patients (Table 2).
Molecular Typing of Bacterial Isolates
The defined cgMLST gene set consisted of a total of 2,831 core and 1,017 accessory targets. Of 77 sequenced isolates, 59 isolates were confirmed as C. sakazakii. These isolates had on average 99.4% of good core genome targets (97.7% to 99.9%) (18) and revealed in total 17 different sequence types (STs) (Table 3).
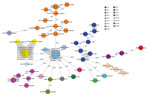
Core genome comparison of 59 C. sakazakii isolates and the 3 reference strains revealed an average allelic difference of 2,402 and a maximum allelic difference of 2,724 (Figure 2). Isolates clustered in the minimum-spanning tree to their respective MLST. Eight isolates belonging to ST1 included 2 stool isolates from neonates with a common epidemiologic link in Austria in 2009; these 8 isolates showed 1 allelic difference and were most closely related (203 alleles difference) to reference ATCC BAA-894, an isolate collected from powdered formula in the United States in 2001. That outbreak affected 2 neonates with necrotizing enterocolitis (both male, age 10 days and 12 days) hospitalized in the same neonatal intensive care unit.
Twelve isolates belonged to ST4, of which 3 were confirmed isolates from infants. Two infant isolates belonged to an outbreak cluster with a common epidemiologic link detected in Austria in 2016; these isolates shared the same cgMLST profile and showed a maximum of 47 allelic differences to the historical reference strain NCTC 8155 (from milk, United Kingdom, 1950). This outbreak again affected 2 neonates (neonate A: female, age 22 days, positive blood culture, fatal outcome; neonate B: male, age 16 days, positive respiratory tract specimen) hospitalized in the neonatal intensive care unit of another hospital in Austria. The third infant isolate was a 2017 ST4 isolate from a case in Austria with a fatal outcome and was most closely related (302 allelic differences) to a historical strain from Denmark isolated in 2003.
Six clinical isolates assigned to ST8 consisted of 2 historical human isolates from Canada (date of isolation unknown). These 6 isolates shared the identical core genome profile and had 1 allelic difference to reference strain ATCC29544 (from an infant, United States, 1970).
Nine human isolates assigned to ST21 comprised a historical outbreak cluster from France collected during 2010–2016. The outbreak included 3 female patients (mean age 62 years) and 5 male patients (mean age 68 years); initial specimens were abscess material from the digestive tract (n = 1), ascites fluid (n = 1), respiratory tract specimens (n = 2), and rectal swab specimens (n = 4). Eight of these 9 isolates showed the same core genome genes, and 1 yielded 1 allelic difference.
All 10 isolates assigned to ST155 belonged to a historical outbreak among infants in Belgium during 1997–1998; the isolates originated from blood cultures (n = 2), stool specimens (n = 2), rectal swab specimens (n = 4), and respiratory tract specimens (n = 2). The first positive sample was collected in November 1997; the remaining 9 specimens were obtained during August–September 1998. Eight of the 10 isolates shared the same cgMLST profile, and 2 had 1 allelic difference.
In total, 27 of 38 historical isolates were most closely related (<1 allelic difference) to other historical isolates; 11 were singletons. All 21 isolates collected in 2017 were singletons, and no close relatedness was evident (>100 allelic differences) between historical isolates and isolates from 2017.
In Vitro and In Silico Antimicrobial Resistance Analysis
In vitro susceptibility testing of 21 human C. sakazakii isolates from 2017 revealed 20 C. sakazakii isolates that were susceptible to all 14 tested antibiotics (Appendix Table 2). One isolate was resistant to ampicillin, cefotaxime, gentamicin (intermediate), moxifloxacin, and trimethoprim/sulfamethoxazole.
Of 21 C. sakazakii isolates, 12 isolates carried the efflux genes emrB, msbA, patA, regulatory systems modulating antibiotic efflux CRP, marA, emrR, marR, H-NS, antibiotic target protection gene msrB, and the determinant of fosfomycin resistance glpT. Seven isolates had in addition the antibiotic protection gene vgaC. One isolate, had also the efflux gene norB, the antibiotic inactivation gene fosX, and the antibiotic target alteration gene mprF. One isolate had the additional antibiotic inactivation genes aac(6')-Ib-cr, aadA16, aadA2, ant(2'')-Ia, arr-3, catB3, CTX-M-9, OXA-1, the antibiotic target protection gene qnrA1, and the antibiotic target replacement gene sul1.
The presence of variant ampC was confirmed for all 21 isolates. Seventeen isolates harbored CSA-2, and 4 isolates harbored CSA-1 (Appendix Table 2).
The aim of our 2017 C. sakazakii study was to assess the occurrence of this opportunistic pathogen in countries of Europe, characterize the isolates, and recognize possible multinational outbreaks. Our finding that only 59 of 77 presumptive C. sakazakii isolates had the species-identification C. sakazakii confirmed at the central study center shows that correct identification of Cronobacter spp. is still a challenge for many routine laboratories.
The prevalence of reported C. sakazakii cases was low, with only 11 (45.8%) of 24 participating countries submitting C. sakazakii isolates. Clinical isolates from 2017 showed high genetic diversity, indicating that neither multinational nor national outbreaks occurred in 2017 in the 24 countries studied. However, characterization of the historical isolates obtained during this study confirmed occurrence of 4 previously unpublished historical outbreaks: 2 outbreaks from 2009 and 2016 in Austria, 1 from Belgium during 1997–1998, and 1 from France during 2010–2016. Hospitals affected by nosocomial C. sakazakii outbreaks might still be reluctant to publish possibly food-related outbreaks or nosocomial infections, especially in the case of affected infants and particularly in the case of related fatalities.
Strain typing using classical MLST identified a total of 17 STs among 59 sequenced C. sakazakii isolates. Our addition of a new ad hoc cgMLST scheme consisting of 2,831 core target genes provides more discriminative power for outbreak investigation and source tracking than the standard 7-loci MLST scheme.
The dominant STs found among our clinical C. sakazakii isolates from 2017 were ST4, ST17, ST1, and ST40, a distribution consistent with results from other studies (1). The medical literature often links C. sakazakii ST4 with powdered infant formula–associated outbreaks in infants (3). In our study, the sole strain (7750-17) affecting an infant (a 3-month-old baby boy who died) was ST4, isolated from a blood culture.
Antibiotic treatment is essential in the care of a patient with a confirmed Cronobacter infection. The traditional antibiotic regimen for Cronobacter spp. was ampicillin in combination with either gentamicin or chloramphenicol. In view of claimed resistance to ampicillin and most first- and second-generation cephalosporins, it has been suggested that carbapenems or third-generation cephalosporins be used with an aminoglycoside or trimethoprim/sulfamethoxazole (24). In our study, antimicrobial resistance testing showed susceptibility to all tested antibiotics for 20 of 21 human isolates from 2017. In comparison to other members of the family Enterobacteriaceae, Cronobacter strains seem to be more susceptible against so-called “key access antibiotics” of the World Health Organization’s Model List of Essential Medicines (25), such as ampicillin, aminoglycosides, chloramphenicol, and third-generation cephalosporins (the last is included in the List of Essential Medicines only for specific, limited indications) (26). For all isolates, we confirmed the presence of 1 of 4 tested ampC β-lactamase variants, which confer phenotypic resistance exclusively to first-generation cephalosporins (e.g., cephalothin) but not to ampicillin (23). A few studies have reported Cronobacter isolates conferring multidrug resistance (26), a phenomenon observed in our study only for 1 strain from Slovenia.
Correct species identification within the Cronobacter group was a major challenge for 7 of 11 participating laboratories. This identification problem is consistent with numerous misidentifications reported in the literature (27,28). The discrepancies in correct Cronobacter spp. identification on a genus and species level between the study center in Austria and the primary testing laboratories using MALDI-TOF MS is probably attributable to outdated databases used by primary testing laboratories. Nevertheless, our study showed that the overall MALDI-TOF MS performance for Cronobacter spp. identification on species level is insufficient and misleading. The databases contained data for C. sakazakii only, and therefore all 7 species of the genus Cronobacter were identified as C. sakazakii. In addition, although a database comment indicated that Cronobacter could only be identified on the genus level, the MALDI-TOF MS result simulated the highest identification score for C. sakazakii. This shortfall should be corrected by an update of the MALDI-TOF MS databases to enable accurate Cronobacter identification at the species level. In comparison, WGS-based species identification represents a major improvement to conventional identification methods and MALDI-TOF MS (29). Therefore, we recommend the use of WGS-based identification tools and databases for identification of species within the Cronobacter group.
Adults were the main affected age group in our study. All but 2 of the isolates from 2017 originated from adults. This finding confirms the results from previous recent studies (14,30) and contradicts statements in numerous medical textbooks, postulating that infants are more often affected than adults (8,31–33).
Our study has some limitations. Lack of information (e.g., detailed epidemiologic and clinical patient data) and misidentification on genus and species levels might have played a role in underestimating the real prevalence rate; 13 of the 24 participating countries did not find or did not submit C. sakazakii isolates.
In conclusion, this C. sakazakii study in Europe revealed a high strain diversity, which points to highly diverse infection sources and an absence of national or multinational outbreaks in 2017. Correct identification of C. sakazakii still poses a diagnostic challenge to many laboratories, and the use of such imperfect detection systems might explain the low prevalence of reported clinical C. sakazakii isolates found in this study. WGS data must be used for accurate species identification and high-resolution strain typing. We recommend the inclusion of C. sakazakii as a notifiable organism by public health authorities.
Ms. Lepuschitz studied molecular biology with a focus on molecular medicine and is currently a PhD student focusing on infectious diseases at the Vienna University of Technology and the Austrian Agency for Health and Food Safety, Vienna, Austria. Her primary research interests include the characterization and epidemiology of clinical, foodborne, and environmental pathogens.
Acknowledgments
We thank the following persons for their willingness to provide isolates and information to the national study coordinators: Charlotte Nielsen Agergaard, John Bingham, Nadine Botteldoorn, Carolyn Cameron, Urska Dermota, Harald Dirschmid, Astrid Dopita, Stefan Doppler, Gebhard Feierl, Manfred Fille, Sonja Fischelschweiger, J. Gigi, Maja Gošnjak, Nils Grude, Renée Haunold, Iren Høyland Löhr, Heidrun Kerschner, Uwe König, Helena Ramos, Tanja Vrabič, Slavica Lorencic Robnik, Tatjana Rupel, Helene Marchandin, Helga Masoud, Louise O’Connor, Renata Ocvirk, Christian Petternel, Catherine Potvliege, Christian Salgård Jensen, Gunnar Skov Simonsen, Bettina Titz, Herwig Tomantschger, Martin Tötsch, Svetlana Ugarcina Perovic, Birgit Willinger, and Markus Winkler.
This study was initiated and partially funded by ESCMID Food- and Water-borne Infections Study Group.
The authors have no conflict of interests to declare.
The institutional review board of the city of Vienna studied the protocol and decided on July 28, 2016, under EK 16-161-VK-NZ that the study did not require formal ethics review.
Members of the EUCRONI study group: F. Allerberger, Austrian Agency for Health and Food Safety, Vienna, Austria; A. Tambic Andrasevic, University Hospital for Infectious Diseases, Zagreb, Croatia; A. Balode, Pauls Stradiņš Clinical University Hospital, Riga, Latvia; F. Barbut, Hôpital Saint-Antoine, Paris, France; Irina Codita, Carol Davila University of Medicine and Pharmacy, Bucharest, Romania; M. Cormican, School of Medicine National University of Ireland Galway, Galway, Ireland; C. Ferguson, Victoria Hospital, Kirkcaldy, Scotland, United Kingdom; P. Heczko, Jagiellonian University Medical College, Kraków, Poland; O. Holy, Department of Preventive Medicine, Palacky University Olomouc, Czech Republic; T. Kantardjiev, National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria; E.J. Kuijper, Leiden University Medical Center, Leiden, the Netherland; T.M. Leegaard, Akershus University Hospital, Lørenskog, Norway; L.M.V. Peixe, University of Porto, Porto, Portugal; D. Piérard, Department of Laboratory Medicine, Universitair Ziekenhuis Brussel, Belgium; H. Rautelin, Uppsala University, Uppsala, Sweden; M. Rupnik, Nacionalni Laboratorij za Zdravje, Okolje in Hrano, Maribor, Slovenia; K. Schønning, Hvidovre Hospital, Hvidovre, Denmark; R. Stephan, Institute for Food Safety and Hygiene, University of Zurich, Zurich, Switzerland; A. Toniolo, Università degli Studi dell’Insubria, Varese, Italy; T. Tošić, Clinical Center of Serbia, Belgrade, Serbia; S. Valdezate, Instituto de Salud Carlos III., Madrid, Spain; L. von Müller, Christophorus-Kliniken GmbH, Coesfeld, Germany; L. Zerva, Attikon-Hospital, Athens, Greece; and B. Zinieri-Panayide, General Hospital, Paphos, Cyprus.
References
- Forsythe SJ, Dickins B, Jolley KA. Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics. 2014;15:1121. DOIPubMedGoogle Scholar
- Iversen C, Mullane N, McCardell B, Tall BD, Lehner A, Fanning S, et al. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int J Syst Evol Microbiol. 2008;58:1442–7. DOIPubMedGoogle Scholar
- Joseph S, Forsythe SJ. Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections. Emerg Infect Dis. 2011;17:1713–5. DOIPubMedGoogle Scholar
- Kandhai MC, Reij MW, Gorris LG, Guillaume-Gentil O, van Schothorst M. Occurrence of Enterobacter sakazakii in food production environments and households. Lancet. 2004;363:39–40. DOIPubMedGoogle Scholar
- Lehner A, Stephan R. Microbiological, epidemiological, and food safety aspects of Enterobacter sakazakii. J Food Prot. 2004;67:2850–7. DOIPubMedGoogle Scholar
- Iversen C, Forsythe S. Risk profile of Enterobacter sakazakii, an emergent pathogen associated with infant milk formula. Trends Food Sci Technol. 2003;14:443–54. DOIGoogle Scholar
- European Food Safety Agency. Opinion of the Scientific Panel on Biological Hazards (BIOHAZ) related to the microbiological risks in infant formulae and follow-on formulae. European Food Safety Agency Journal. 2004;113:1–35.
- Lund BM. Properties of microorganisms that cause foodborne disease. In: Lund BM, Hunter PR, editors. The microbiological safety of food in healthcare settings. Oxford: Blackwell Publishing; 2008. p. 12–233.
- Forsythe SJ. Updates on the Cronobacter genus. Annu Rev Food Sci Technol. 2018;9:23–44. DOIPubMedGoogle Scholar
- World Health Organization and Food and Agriculture Organization of the United Nations. Enterobacter sakazakii (Cronobacter spp.) in powdered follow-up formula. Meeting report, Microbiology Risk Assessment Series, No. 15. Geneva: World Health Organization; 2008 [cited 2018 Oct 1. http://www.who.int/foodsafety/publications/micro/MRA_followup.pdf
- Losio MN, Pavoni E, Finazzi G, Agostoni C, Daminelli P, Dalzini E, et al. Preparation of powdered infant formula: could product’s safety be improved? J Pediatr Gastroenterol Nutr. 2018;67:543–6. DOIPubMedGoogle Scholar
- Chaves CEV, Brandão MLL, Lacerda MLGG, Rocha CABC, Leone de Oliveira SMDV, Parpinelli TC, et al. Fatal Cronobacter sakazakii sequence type 494 meningitis in a newborn, Brazil. Emerg Infect Dis. 2018;24:1948–50. DOIPubMedGoogle Scholar
- Drudy D, Mullane NR, Quinn T, Wall PG, Fanning S. Enterobacter sakazakii: an emerging pathogen in powdered infant formula. Clin Infect Dis. 2006;42:996–1002. DOIPubMedGoogle Scholar
- Alsonosi A, Hariri S, Kajsík M, Oriešková M, Hanulík V, Röderová M, et al. The speciation and genotyping of Cronobacter isolates from hospitalised patients. Eur J Clin Microbiol Infect Dis. 2015;34:1979–88. DOIPubMedGoogle Scholar
- Patrick ME, Mahon BE, Greene SA, Rounds J, Cronquist A, Wymore K, et al. Incidence of Cronobacter spp. infections, United States, 2003-2009. Emerg Infect Dis. 2014;20:1520–3. DOIPubMedGoogle Scholar
- Lepuschitz S, Sorschag S, Springer B, Allerberger F, Ruppitsch W. Draft genome sequence of carbapenemase-producing Serratia marcescens isolated from a patient with chronic obstructive pulmonary disease. Genome Announc. 2017;5:e01288–17. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, et al. Defining and evaluating a core genome MLST scheme for whole genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol. 2015;53:2869–76. DOIPubMedGoogle Scholar
- Kucerova E, Clifton SW, Xia XQ, Long F, Porwollik S, Fulton L, et al. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One. 2010;5:e9556. DOIPubMedGoogle Scholar
- Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–31. DOIPubMedGoogle Scholar
- Jolley KA, Bliss CM, Bennett JS, Bratcher HB, Brehony C, Colles FM, et al. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology. 2012;158:1005–15. DOIPubMedGoogle Scholar
- Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45(D1):D566–73. DOIPubMedGoogle Scholar
- Müller A, Hächler H, Stephan R, Lehner A. Presence of AmpC beta-lactamases, CSA-1, CSA-2, CMA-1, and CMA-2 conferring an unusual resistance phenotype in Cronobacter sakazakii and Cronobacter malonaticus. Microb Drug Resist. 2014;20:275–80. DOIPubMedGoogle Scholar
- Lai KK. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore). 2001;80:113–22. DOIPubMedGoogle Scholar
- World Health Organization. The selection and use of essential medicines. Report of the WHO Expert Committee, 2017 (including the 20th WHO Model List of Essential Medicines and the 6th WHO Model List of Essential Medicines for Children). WHO Technical Report Series, no. 1006. Geneva: The Organization; 2017 [cited 2018 Oct 1]. http://apps.who.int/medicinedocs/documents/s23371en/s23371en.pdf
- Shi L, Liang Q, Zhan Z, Feng J, Zhao Y, Chen Y, et al. Co-occurrence of 3 different resistance plasmids in a multi-drug resistant Cronobacter sakazakii isolate causing neonatal infections. Virulence. 2018;9:110–20. DOIPubMedGoogle Scholar
- Blažková M, Javůrková B, Vlach J, Göselová S, Karamonová L, Ogrodzki P, et al. Diversity of O antigens within the genus Cronobacter: from disorder to order. Appl Environ Microbiol. 2015;81:5574–82. DOIPubMedGoogle Scholar
- Jackson EE, Flores JP, Fernández-Escartín E, Forsythe SJ. Reevaluation of a suspected Cronobacter sakazakii outbreak in Mexico. J Food Prot. 2015;78:1191–6. DOIPubMedGoogle Scholar
- Yan Q, Fanning S. Strategies for the identification and tracking of cronobacter species: an opportunistic pathogen of concern to neonatal health. Front Pediatr. 2015;3:38. DOIPubMedGoogle Scholar
- Holý O, Forsythe S. Cronobacter spp. as emerging causes of healthcare-associated infection. J Hosp Infect. 2014;86:169–77. DOIPubMedGoogle Scholar
- Allerberger F, Pichler J, Öhlinger R, Gelpi E, Budka H. Nahrungsmittelbedingte Infektionskrankheiten und Intoxikationen. In: Ledochowski M, editor. Klinische Ernährungsmedizin. Vienna: Springer-Verlag; 2010. p. 347–408.
- Barer MR, Swann A. Klebsiella, Enterobacter, Proteus and other enterobacteria. In: Barer MR, Irving W, Swann A, Perera N, editors. Medial microbiology—a guide to microbial infections: pathogenesis, immunity, laboratory investigation and control. 19th edition. Oxford: Elsevier Inc.; 2018. p. 198–204.
- Jenkins C, Rentenaar RJ, Landraid L, Brisse S. Enterbacteriaceae. In: Cophen J, Powderly WG, Opal SM, editors. Mandell, Douglas and Bennett’s principles and practice of infectious diseases. 8th edition. Atlanta: Elsevier Inc.; 2017. p. 1564–78.
Figures
Tables
Cite This ArticleOriginal Publication Date: February 07, 2019
1Members of the EUCRONI study group are listed at the end of this article.
Table of Contents – Volume 25, Number 3—March 2019
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Sarah Lepuschitz, Austrian Agency for Health and Food Safety, Institute for Medical Microbiology and Hygiene, Waehringerstraße 25a, 1090 Vienna, Austria
Top