Volume 25, Number 9—September 2019
Research
Theileria orientalis Ikeda Genotype in Cattle, Virginia, USA
Figure 4
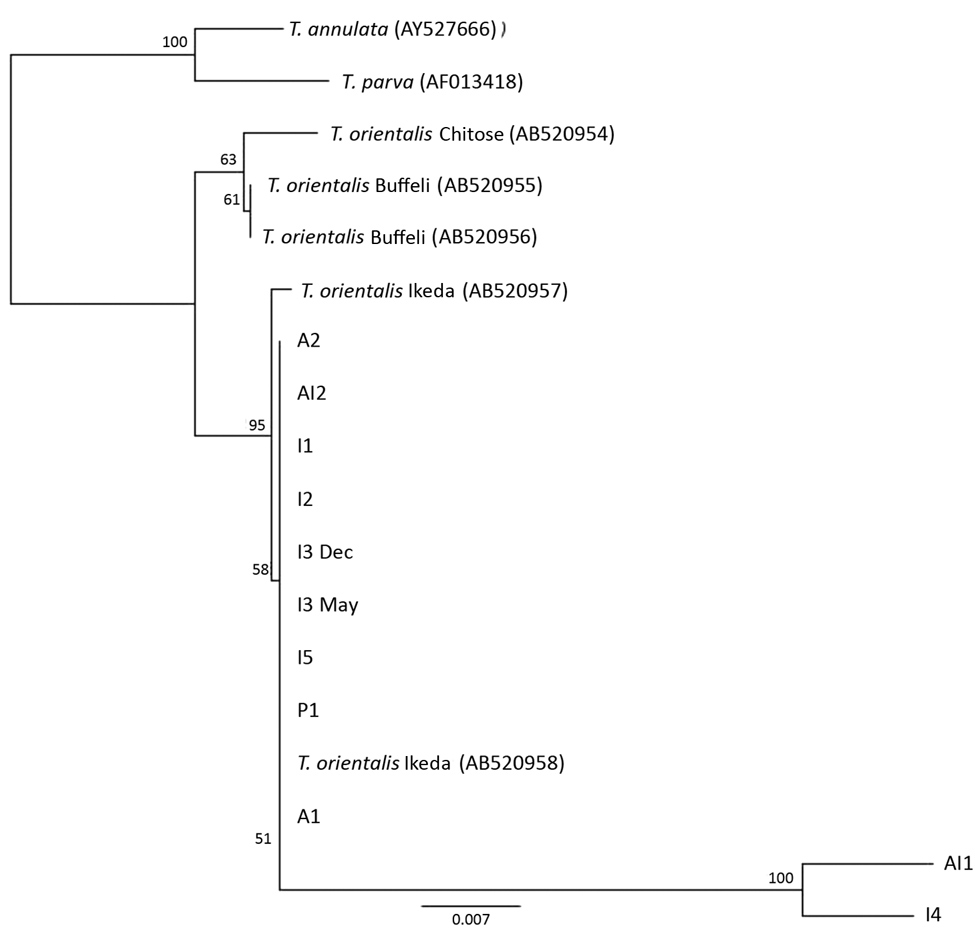
Figure 4. Phylogenetic tree showing small subunit rDNA gene sequences for Theileria species. Representative samples from cattle in 6 herds in Virginia, USA, cluster with the reference sequences for the Ikeda genotype. The next most closely related branch is composed of T. orientalis Chitose genotype and then T. orientalis Buffeli genotype. The outgroup is composed of a single reference sequence each for T. parva and T. annulata. Small subunit rDNA sequences for I4 and Al1 were of low quality (6.4% and 3.9%, respectively). Numbers along branches are bootstrap values. Scale bar indicates nucleotide substitutions per site.
Page created: August 21, 2019
Page updated: August 21, 2019
Page reviewed: August 21, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.