Volume 26, Number 12—December 2020
Dispatch
Hypervirulent Klebsiella pneumoniae as Unexpected Cause of Fatal Outbreak in Captive Marmosets, Brazil
Cite This Article
Citation for Media
Abstract
After the sudden death of captive marmosets in São Paulo, Brazil, we conducted a histologic and microbiologic study. We found hyperacute septicemia caused by hypermucoviscous sequence type 86 K2 Klebsiella pneumoniae. We implemented prophylactic antimicrobial therapy, selected dedicated staff for marmoset interactions, and sanitized the animals’ fruit to successfully control this outbreak.
Klebsiella pneumoniae is an opportunistic bacteria that is a normal part of the nasopharyngeal and gastrointestinal tract microbiome of humans and animals (1). The hypermucoviscous variant of K. pneumoniae (hvKp), initially described in Southeast Asia, has emerged as a pathogen affecting young and healthy persons worldwide (2). The development of prominent polysaccharide capsules associated with capsular serotypes K1 or K2 have been reported as the major virulence determinants for human hvKp in liver abscesses, perhaps because it seems to protect the bacteria from phagocytosis and prevents destruction by bactericidal serum factors (2).
K. pneumoniae strains have also been associated with a variety of diseases in animals, especially in Old World (Africa, Asia, and Europe) and New World (Oceana, North America, and South America) nonhuman primates (3–5). Sudden death or various clinical signs, including anorexia, prostration, fever, cough, dyspnea, mucopurulent discharge, meningitis, pneumonia, peritonitis, and sepsis are strongly associated with sporadic infections of K. pneumoniae in common marmosets research colonies (5,6).
Despite the well-recognized zoonotic importance of hvKp and the public health risk of emerging multidrug-resistant strains (7–9), information is incomplete about the genotypic and phenotypic characterization of the etiologic agent essential to adequately diagnose and treat this pathogen in captive and wild nonhuman primates. The aim of this study was to report an epizootic among common marmosets in a wildlife rehabilitation center in Brazil and to describe the serotype, sequence typing, virulence properties, and resistance profile of the K. pneumoniae strains involved.
On February 10–11, 2019, a total of 11 captive marmosets (8 Callithrix penicillata, 2 C. jacchus, and 1 hybrid) died suddenly without clinical signs of disease. All of the animals were maintained in Parque Ecológico do Tietê, located in São Paulo municipality, São Paulo, Brazil, which is a center for receiving, rehabilitating, and referring wildlife. All animals had been in captivity 123–399 days. No new animals had been introduced into the cages in the previous 25 days. Each necropsy was performed <24 h after death in accordance with the Brazil Ministry of Health’s guide for surveillance of epizootics in nonhuman primates (10). Tissue samples were preserved in phosphate-buffered formalin 10% and processed for routine histopathology and for 12 hours in refrigeration for microbiologic and molecular analyses. This study was approved by the Ethics Committee for the Use of Animals (CEUA) of Adolfo Lutz Institute, Brazil (protocol no. 11/2016), SISBIO registration no. 50551 for the manipulation of wildlife material, and SISGEN registration nos. A1A2A72 and A7EB4B6.
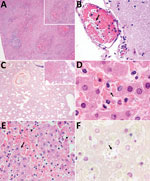
Histologic findings from all of the animals were compatible with hyperacute septicemia. Multiple sections of liver, spleen, and adrenal tissue were similarly affected by suppurative and necrotizing multifocal lesions associated with intrahistiocytic gram-negative bacteria, 1–2-μm long. We also observed numerous intravascular gram-negative bacilli in samples from the liver (10 of 10 samples; the sample from 1 marmoset was excluded because the animal showed severe postmortem autolysis), cerebrum (8/10), lungs (3/10), heart (1/7), intestines (1/7), thymus (1/2), and skeletal muscle (1/1). Other relevant microscopic findings from different tissues are summarized in Table 1 and Figure 1. We found no microscopic alterations in the analyzed fragment samples from the stomach, tongue, testis, thymus, skin, uterus, or lymph nodes.
Pure cultures of K. pneumoniae, positive for the string test (11), were recovered from the brain and liver in 8 different animals. The representative isolate P04 displayed susceptibility to all the antimicrobial agents we evaluated using the broth microdilution methodology with Sensititre plate (Thermo Fisher Scientific, https://www.thermofisher.com), according to the manufacturer’s instructions (Table 2). We screened for K. pneumoniae in environmental samples of water from a lake near the primate cages by filtration and in drag swabs from their cages by direct growth on MacConkey agar plates. Although we recovered K. pneumoniae isolates from both samples, all of them were negative in the string test. We subjected all isolates identified as K. pneumoniae to DNA macro-restriction by using 30 U of XbaI enzyme followed by pulsed-field gel electrophoresis (PFGE) (https://www.cdc.gov/pulsenet/pathogens/pfge.html). PFGE results showed the same restriction profile among the isolates recovered from the dead animals; however, environmental isolates clustered apart from the invasive isolates (Figure 2).
We subjected the P04 strain to whole genome sequencing using the Thermo Fisher Ion Torrent S5 platform, resulting in 1,049,163 reads; the de novo assembled genome comprised 5,358,608 bps grouped in 62 contigs, with an average coverage depth of 53. The whole-genome shotgun project reported here was deposited in DDBJ/EMBL/GenBank under accession number SPSP00000000.
We employed online platforms from PubMLST (https://pubmlst.org) to definitively identify species as K. pneumoniae, sequence type (ST) 86, capsular type K2, and multiple virulence genes: mrkABCDFHIJ cluster (mannose-resistant Klebsiella-like type III fimbriae cluster, associated with adhesiveness and fimbrial filament formation to adhere to eukaryotic cells) (12) in the same contig of the kvgAS genes; iroBCD genes (salmochelin) in the same contig with the rmpA gene (regulator of mucoid phenotype A); and rmpA2 gene within the contig along the iucABD with the iutA genes (aerobactin) (http://bigsdb.pasteur.fr). We detected only the constitutive antimicrobial-resistant genes blaSHV-1, oqxAB, and fosA by the in silico analysis (Comprehensive Antibiotic Resistance Database, https://card.mcmaster.ca). Phylogenetic analysis of high quality single-nucleotide polymorphisms (SNPs) built on the CSI Phylogeny 1.4 (Center for Genomic Epidemiology, https://cge.cbs.dtu.dk/services/CSIPhylogeny) showed that the ST86 isolates were closely related and the P04 strain clustered closely with IPEUC340, an isolate recovered in 1975 in France (Appendix). The isolate RJF293, ST374, clustered apart from the ST86 isolates (Appendix Figure).
To contain the spread of hvKp to other animals, metaphylatic therapy with trimethoprim/sulfamethoxazole was implemented by adding the antimicrobial to the water supply of animals during the first 5 days after the fatal cases were identified. In addition, access to the cages was restricted to a dedicated employee, who wore clothing exclusively for accessing the cages for the duration of the outbreak. We also implemented additional steps for sanitizing fruit eaten by the marmosets with 2% sodium hypochlorite for 15 minutes and dedicated space and staff for preparing the marmosets’ meals after the epizootic event.
We report the detection of hypermucoviscous K. pneumoniae ST86 K2 as cause of a sudden fatal outbreak in captive marmosets. Implementing prompt containment measures led to successful control of this outbreak. The burden of hypermucoviscous K. pneumoniae ST86 K2 in unexpected reservoirs, including those in contact with people, deserves further investigation. The emergence of these strains is a concern for human and veterinary health because of the potential for these bacteria to acquire multidrug-resistant genes, their capacity to persist in the environment and to infect a wide range of hosts, and the unavailability of vaccines against these strains for humans and animals (13). The expansion of this emerging pathogen among different reservoirs should be carefully surveilled, because the relationship between hypervirulent and multidrug-resistant strains is narrowing.
Acknowledgments
We thank the team of curators of the Institut Pasteur MLST and whole genome MLST databases for curating the data and making them publicly available and all contributors from the Center of Pathology at Adolfo Lutz Institute in São Paulo for routine sample processing.
Dr. Guerra is a veterinary pathologist and scientific researcher at the Center of Pathology at Adolfo Lutz Institute in São Paulo, Brazil. Her research focuses on the comparative pathology of emerging and reemerging infectious diseases in the context of an integrated One Health approach.
References
- Bueno MG, Iovine RO, Torres LN, Catão-Dias JL, Pissinatti A, Kierulff MC, et al. Pneumonia and bacteremia in a golden-headed lion tamarin (Leontopithecus chrysomelas) caused by Klebsiella pneumoniae subsp. pneumoniae during a translocation program of free-ranging animals in Brazil. J Vet Diagn Invest. 2015;27:387–91. DOIPubMedGoogle Scholar
- Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32:
e00001-19 . DOIPubMedGoogle Scholar - Fox JG, Rohovsky MW. Meningitis caused by Klebsiella spp in two rhesus monkeys. J Am Vet Med Assoc. 1975;167:634–6.PubMedGoogle Scholar
- Gonzalo A, Montoya E. Klebsiella pneumoniae infection in a New World nonhuman primate center. Laboratory Primate Newsletter 1991;30:13–20 [cited 2019 Oct 20]. https://www.brown.edu/Research/Primate/lpn30-2.html#kleb
- Pisharath HR, Cooper TK, Brice AK, Cianciolo RE, Pistorio AL, Wachtman LM, et al. Septicemia and peritonitis in a colony of common marmosets (Callithrix jacchus) secondary to Klebsiella pneumoniae infection. Contemp Top Lab Anim Sci. 2005;44:35–7.PubMedGoogle Scholar
- Kindlovits A, Kindlovits L. Clinic and therapeutics in neotropical primates [in Portuguese]. 2nd ed. Rio de Janeiro: L.F. Livros; 2009.
- Wu F, Ying Y, Yin M, Jiang Y, Wu C, Qian C, et al. Molecular characterization of a multidrug-resistant Klebsiella pneumoniae strain R46 isolated from a rabbit. Int J Genomics. 2019;2019:
5459190 . DOIPubMedGoogle Scholar - Osman KM, Hassan HM, Orabi A, Abdelhafez AST. Phenotypic, antimicrobial susceptibility profile and virulence factors of Klebsiella pneumoniae isolated from buffalo and cow mastitic milk. Pathog Glob Health. 2014;108:191–9. DOIPubMedGoogle Scholar
- Boszczowski I, Salomão MC, Moura ML, Freire MP, Guimarães T, Cury AP, et al. Multidrug-resistant Klebsiella pneumoniae: genetic diversity, mechanisms of resistance to polymyxins and clinical outcomes in a tertiary teaching hospital in Brazil. Rev Inst Med Trop São Paulo. 2019;61:
e29 . DOIPubMedGoogle Scholar - Brasil Ministério da Saúde. Guide for surveillance of epizootics in nonhuman primates and entomology applied to yellow fever surveillance [in Portuguese]. 2nd ed. Brasília: Brazil Ministry of Health; 2014.
- Siu LK, Yeh K-M, Lin J-C, Fung C-P, Chang F-Y. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12:881–7. DOIPubMedGoogle Scholar
- Gerlach GF, Clegg S, Allen BL. Identification and characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae. J Bacteriol. 1989;171:1262–70. DOIPubMedGoogle Scholar
- Cox BL, Schiffer H, Dagget G Jr, Beierschmitt A, Sithole F, Lee E, et al. Resistance of Klebsiella pneumoniae to the innate immune system of African green monkeys. Vet Microbiol. 2015;176:134–42. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: November 08, 2020
Table of Contents – Volume 26, Number 12—December 2020
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Carlos Henrique Camargo, Instituto Adolfo Lutz, Centro de Bacteriologia, Núcleo de Doenças Entéricas e Infecções por Patógenos Especiais, Avenida Dr. Arnaldo, 351–9° Andar, Pacaembú, São Paulo, Brazil
Top