Volume 26, Number 12—December 2020
Research
Equine-Like H3 Avian Influenza Viruses in Wild Birds, Chile
Figure 4
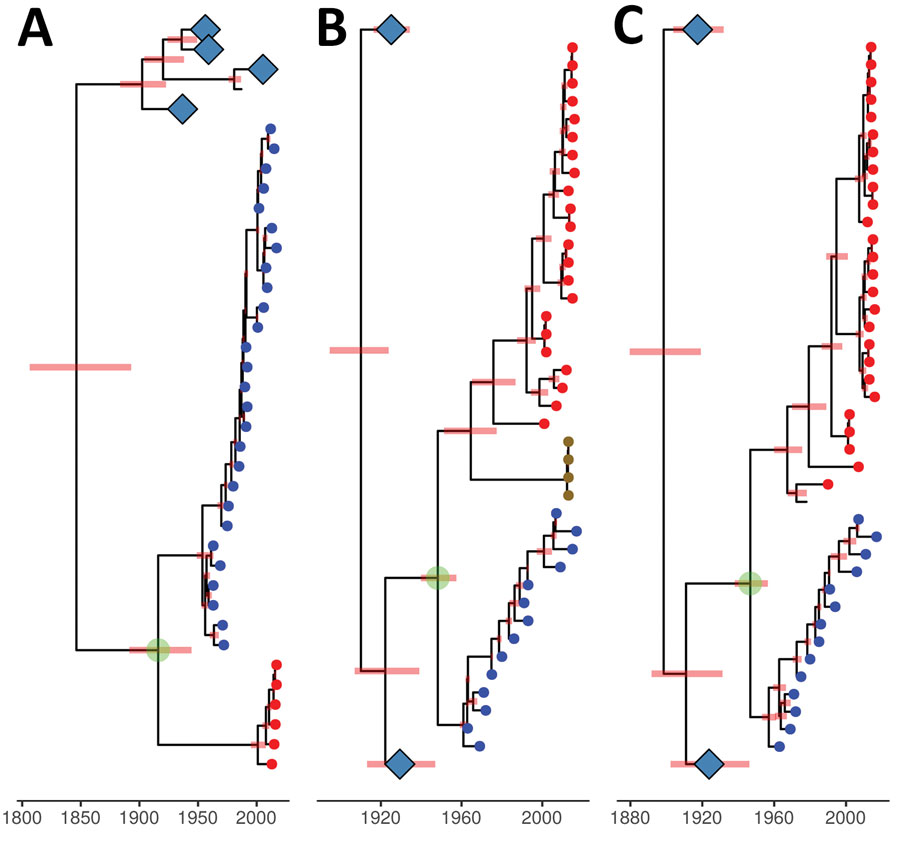
Figure 4. Genetic relationship and divergence date between avian influenza viruses (AIVs) from Chile/South America, the 1963 H3N8 equine-like influenza virus (EIV), and AIVs from other locations. A) Maximum clade credibility tree for the H3 gene segment. B) Maximum clade credibility tree for the polymerase acidic gene segment. AIV samples for penguins from Antarctica are represented by gold circular nodes. C) Maximum clade credibility tree for the nucleoprotein gene segment. Viruses were dated by using Bayesian Markov chain Monte Carlo analysis. Position of tips represent sampled virus for the years they were sampled. H3N8 EIV sequences are represented by blue circular nodes, AIVs from Chile/South America are represented by red circular nodes, and avian sequences from other locations are unlabeled. Internal nodes are reconstructed common ancestors, and pink bars represent 95% credible intervals on their date. Large clades of avian sequences from other locations are collapsed on their common ancestors and represented by light blue diamonds. The common ancestor between most AIVs from South America and the 1963 H3N8 EIV is highlighted by a green circle. Time scale bar indicates years. .
1These authors contributed equally to this article.