Volume 26, Number 4—April 2020
Dispatch
Crimean-Congo Hemorrhagic Fever Virus in Humans and Livestock, Pakistan, 2015–2017
Cite This Article
Citation for Media
Abstract
We detected Crimean-Congo hemorrhagic fever virus infections in 4 provinces of Pakistan during 2017–2018. Overall, seroprevalence was 2.7% in humans and 36.2% in domestic livestock. Antibody prevalence in humans was highest in rural areas, where increased contact with animals is likely.
Crimean-Congo hemorrhagic fever (CCHF) is caused by CCHF virus (CCHFV), an emerging zoonotic virus belonging to the order Bunyavirales within the family Nairoviridae. The virus is maintained through a tick–vertebrate transmission cycle (1); the primary vectors are ticks from the genus Hyalomma (2,3). Wild and domestic mammals, including livestock species such as sheep, goats, and cattle, are amplifying hosts (2). CCHFV is listed as a high-priority zoonotic pathogen of humans in the World Health Organization Research and Development Blueprint (https://www.who.int/blueprint/priority-diseases) because of its potential to cause a public health emergency and the absence of specific treatment and vaccines.
Most human infections occur through the bite of infected ticks. Blood and other bodily fluids of infected animals represent an additional source for human infections. In humans, CCHF is manifested by fever, headache, vomiting, diarrhea, and muscular pain; bleeding diathesis with multiorgan dysfunction is seen in severe cases (4–6). CCHFV is endemic over a wide geographic area, spanning from western Asia to southern Europe and over most of Africa (2). Since the earliest identified CCHF case in 1976 (7), several outbreaks of CCHFV infection have been reported from Pakistan. Although Pakistan has the fourth highest number of human cases in Asia (2), no comprehensive surveillance study has been conducted to determine the disease prevalence in human and animal populations of Pakistan. Therefore, we determined the countrywide risk for CCHFV infection by detecting the virus and antibodies in livestock, ticks, and humans.
During 2017–2018, we tested 3,710 serum samples from 1,872 humans and 1,838 domestic animals (311 buffaloes, 480 camels, 183 cattle, 440 goats, and 424 sheep) for antibodies against CCHFV (Appendix). We also screened 98 blood plasma samples (24 from goats, 28 from buffalo, and 46 from cows) and 774 ticks (509 Hyalomma spp., 134 Rhipicephalus spp., 77 Haemaphysalis spp., and 54 Rhipicephalus [Boophilus] spp.), sampled from livestock in Punjab Province, for CCHFV antigen by commercial ELISA (VectoCrimea-CHF-antigen ELISA; Vector-Best, https://vector-best.ru).
We found a total of 51 (2.7%) human samples to be positive for CCHF antibodies by using a 2-step approach, ELISA and confirmatory testing by immunofluorescence assay (Figure 1; Appendix). We observed significantly higher than average prevalence (p<0.01) among samples from Balochistan (5.7%, 95% CI 3.4%–9.3%) and the lowest prevalence among those from Sindh (1.1%, 95% CI 0.5%–2.3%). Samples from Balochistan were almost 6 times (odds ratio [OR] 5.6, CI 2.0–18.0) more likely to test positive than those from Sindh. Seroprevalence increased uniformly with age; we saw the highest level of CCHFV antibodies in persons >65 years of age (Table 1). Of the 51 positive samples, 28 (2.7%, 95% CI 1.8%–3.8%) were from female and 23 (2.8%, 95% CI 1.9%–4.2%) from male participants. We observed significantly higher (p<0.01) seroprevalence among livestock farmers (3.2%, 95% CI 2.4%–4.2%) compared with the general population (0.6%, 95% CI 0.1%–2.3%).
Of the 1,838 animals, 666 (36.2%) were positive for CCHF by a commercial ELISA (ID Vet, https://www.id-vet.com). The prevalence of CCHFV antibodies was significantly higher (p<0.01) among camels (56.7%, 95% CI 52.1%–61.2%) than among cattle (44.3%, 95% CI 36.9%–51.8%), sheep (32.6%, 95% CI 28.1%–37.2%), buffalo (29.6%, 95% CI 24.6%–35%), and goats (18.9%, 95% CI 15.3%–22.8%) (Appendix Tables 1–5). Camels were almost 6 times (OR 5.6) more likely to be positive than other species. As we found for humans, we found significantly higher (p<0.01) seroprevalence of CCHFV antibodies among animals from Balochistan (59.3%, 95% CI 54.1%–64.5%) than among animals from the other regions tested (Table 2).
We built a binary logistic regression model to evaluate possible risk factors for CCHFV seropositivity in animals and humans. The final model (Appendix Table 6) at the animal level indicated that the animals with highest risk for being antibody positive are livestock from Balochistan (OR 12.1, 95% CI 7.7–19.1), buffalo (OR 4.4, 95% CI 2.8–6.8), and animals >5 years of age (95% OR 1.3, CI 1.0–1.7). However, the NR2 value of 0.277 and the Hosmer-Lemeshow goodness-of-fit test (χ2 22.005; p = 0.003) indicated that this is a poor model for predicting CCHFV exposure in the sampled livestock population.
In humans, we found the chance of exposure to CCHFV was highest for populations from Balochistan (OR 6.6, 95% CI 2.5–17.5) (Appendix Table 7) and in persons belonging to the herdsman profession (OR 7.3, CI 1.7–30.2). The values of NR2 (0.070) and Hosmer-Lemeshow goodness-of-fit test (χ2 1.490; p = 0.684) indicated that our model is a reasonable model for predicting past exposure to CCHFV in the tested human population.
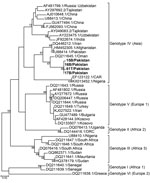
Four plasma samples from buffalo and 4 Rhipicephalus tick samples tested positive for CCHFV antigen by ELISA. Of these 8 positive samples, we confirmed 1 tick (T61) and 3 buffalo samples (15B, 16B, and 17B) through partial amplification and sequencing of the small (S) segment (260 bp). The 4 partial sequences of CCHFV S segments (GenBank accession nos. MN135938–MN135941) were 97%–95% identical to virus sequences found in Afghanistan (accession no. JX908640.1), Iran (accession no. KX096702.1), and Oman (accession no. KY362516.1) and clustered together with genotype IV (Asia) (Figure 2). We obtained full-length sequences of the CCHFV S, medium (M), and large (L) segments (accession nos. MN135942–MN135944) from the tick sample by sequencing on a HiSeq3000 (Illumina, https://www.illumina.com). Phylogenetic trees for the S, M, and L segments showed that the T61 strain clustered with genotype IV (Asia) (Appendix Figures 1–3).
This countrywide study of CCHFV in Pakistan strongly suggests virus circulation in specific geographic regions and suggests CCHFV foci and a potential source of human infections. Detection of the antibodies in domestic livestock species (including sheep, goats, cattle, buffalo, and camels) indicates a potential role of these animals in human infections. Demonstration of the virus in animal blood plasma and tick samples by reverse transcription PCR provides strong evidence of active circulation of CCHFV in Pakistan. Furthermore, genetic characterization of the virus reconfirms the circulation of genotype IV in Pakistan (8). Of interest, we found no Hyalomma tick positive for CCHFV; CCHFV has been reported from Rhipicephalus ticks from Iran and clustered together with strains from Pakistan and Iran, indicating that Rhipicephalus ticks have been naturally infected with closely related virus in the region (9). Our study further confirms the role of Rhipicephalus ticks in CCHFV circulation in the region. We observed higher prevalence of CCHFV antibodies in camels than in animals of other species, indicating the importance of camels in CCHFV ecology in Pakistan.
A high proportion of seropositive humans from Balochistan and Khyber Pakhtunkhwa with a history of exposure to animals is in concordance with earlier reports of CCHF in humans from these areas. The rural economy of Balochistan and Khyber Pakhtunkhwa is based on livestock production, and the increased contact with animals may explain the higher antibody prevalence in humans from these areas. Furthermore, the prevalence of antibodies was significantly higher among herdsmen than among the general population.
In summary, our results indicate the ongoing circulation of CCHFV among animals and humans in some regions of Pakistan. Longitudinal surveys to identify and define the genomic diversity of CCHFV in Pakistan and investigations to explore the exact role of camels in the ecology of this virus would help clarify the risk to the general population and occupational hazards for livestock farmers and veterinarians.
Dr. Zohaib is an assistant professor in the Department of Healthcare Biotechnology, National University of Sciences & Technology, Islamabad, Pakistan. His research interests include the detection and characterization of zoonotic viruses.
Acknowledgments
We thank the Livestock and Dairy Development Departments in Punjab and Balochistan Provinces, Pakistan. We thank the National Virus Resource Center (Wuhan, China) for providing the virus-infected Vero cells (accession no. IVCAS14.0001).
This work was supported by the National Science and Technology major project (2018ZX0101004), the International Cooperation on Key Technologies of Biosafety along the China-Pakistan Economic Corridor (153B42KYSB20170004), the Intergovernmental Special Program of State Key Research and Development Plan from the Ministry of Science and Technology of China (2016YFE0113500), and the Science and Technology Basic Work Program from the Ministry of Science and Technology of China (2013FY113500).
References
- Gargili A, Estrada-Peña A, Spengler JR, Lukashev A, Nuttall PA, Bente DA. The role of ticks in the maintenance and transmission of Crimean-Congo hemorrhagic fever virus: A review of published field and laboratory studies. Antiviral Res. 2017;144:93–119. DOIPubMedGoogle Scholar
- Al-Abri SS, Abaidani IA, Fazlalipour M, Mostafavi E, Leblebicioglu H, Pshenichnaya N, et al. Current status of Crimean-Congo haemorrhagic fever in the World Health Organization Eastern Mediterranean Region: issues, challenges, and future directions. Int J Infect Dis. 2017;58:82–9. DOIPubMedGoogle Scholar
- Bente DA, Forrester NL, Watts DM, McAuley AJ, Whitehouse CA, Bray M. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res. 2013;100:159–89. DOIPubMedGoogle Scholar
- Ergönül O. Crimean-Congo haemorrhagic fever. Lancet Infect Dis. 2006;6:203–14. DOIPubMedGoogle Scholar
- Swanepoel R, Gill DE, Shepherd AJ, Leman PA, Mynhardt JH, Harvey S. The clinical pathology of Crimean-Congo hemorrhagic fever. Rev Infect Dis. 1989;11(Suppl 4):S794–800. DOIPubMedGoogle Scholar
- Whitehouse CA. Crimean-Congo hemorrhagic fever. Antiviral Res. 2004;64:145–60. DOIPubMedGoogle Scholar
- Burney MI, Ghafoor A, Saleen M, Webb PA, Casals J. Nosocomial outbreak of viral hemorrhagic fever caused by Crimean Hemorrhagic fever-Congo virus in Pakistan, January 1976. Am J Trop Med Hyg. 1980;29:941–7. DOIPubMedGoogle Scholar
- Khurshid A, Hassan M, Alam MM, Aamir UB, Rehman L, Sharif S, et al. CCHF virus variants in Pakistan and Afghanistan: Emerging diversity and epidemiology. J Clin Virol. 2015;67:25–30. DOIPubMedGoogle Scholar
- Fakoorziba MR, Naddaf-Sani AA, Moemenbellah-Fard MD, Azizi K, Ahmadnia S, Chinikar S. First phylogenetic analysis of a Crimean-Congo hemorrhagic fever virus genome in naturally infected Rhipicephalus appendiculatus ticks (Acari: Ixodidae). Arch Virol. 2015;160:1197–209. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: March 09, 2020
Table of Contents – Volume 26, Number 4—April 2020
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Zheng-Li Shi, Chinese Academy of Sciences, Key Laboratory of Special Pathogens and Biosafety, Wuhan Institute of Virology, Wuhan 430071, China
Top