Volume 26, Number 5—May 2020
Research Letter
Novel Ehrlichia Strain Infecting Cattle Tick Amblyomma neumanni, Argentina, 2018
Figure
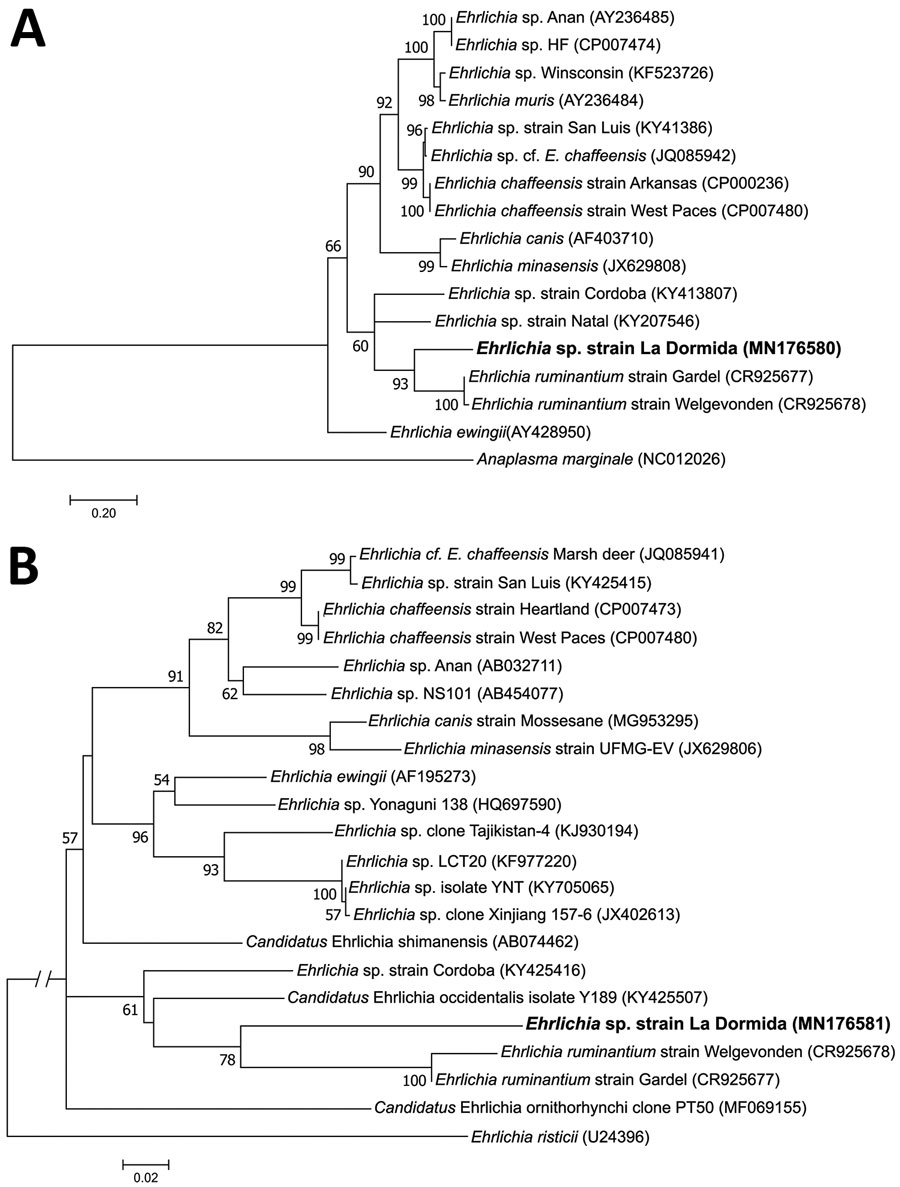
Figure. Maximum-likelihood trees constructed from dsb and groESL sequences of Ehrlichia sp. infecting Amblyomma neumanni ticks in Argentina compared with reference strains. A) Tree constructed by using dsb Ehrlichia sequences of approximately the same length as the sequence identified in this study (341 positions included in the final dataset). B) Tree constructed by using groESL Ehrlichia sequences of approximately the same length as the sequence identified in this study (767 positions included in the final dataset). Phylogenetic trees were constructed by using MEGA 7.0 (https://www.megasoftware.net), and best-fitting substitution models were determined with the Akaike Information Criterion, using the maximum-likelihood model test. Numbers represent bootstrap support generated from 1,000 replications. GenBank accession numbers are shown in parentheses. Boldface indicates the strain identified in this study. Scale bars indicate nucleotide substitutions/site.