Volume 26, Number 6—June 2020
Research Letter
Sabiá Virus–Like Mammarenavirus in Patient with Fatal Hemorrhagic Fever, Brazil, 2020
Figure
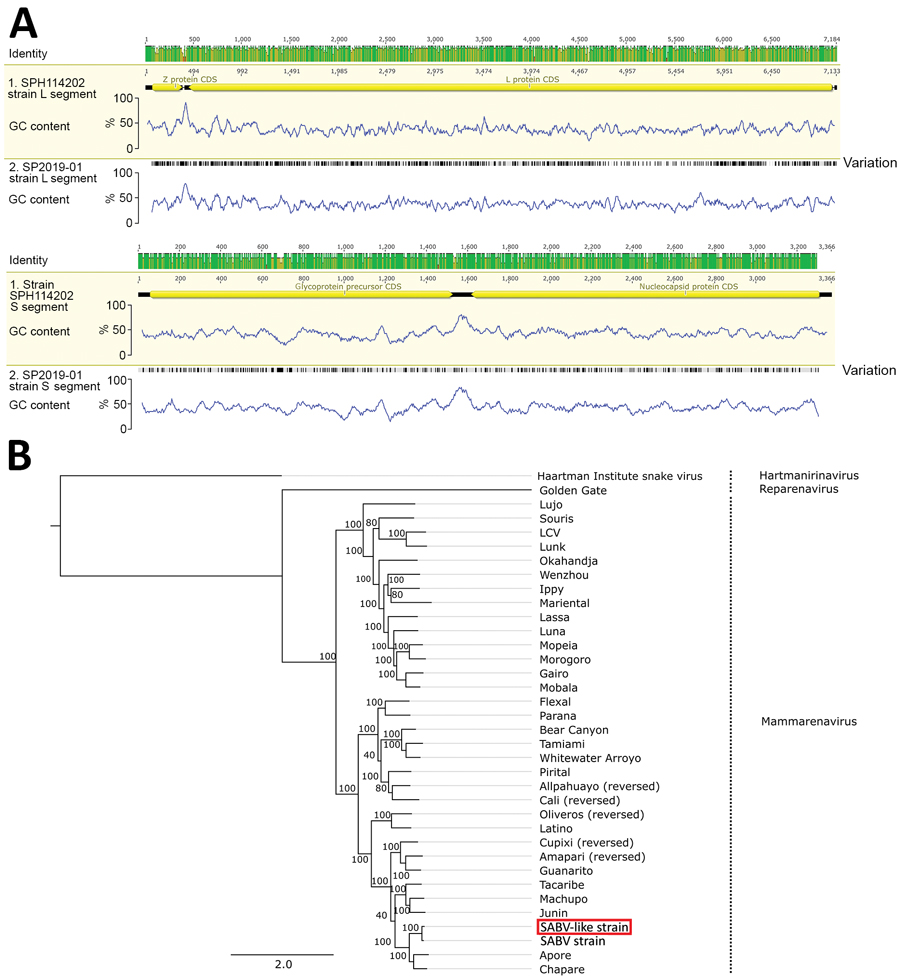
Figure. Genomic and phylogenetic analysis of SABV-like mammarenavirus (SP2019-01) from a patient with fatal hemorrhagic fever, Brazil, 2020. A) Genome plots comparing strain SP2019-01 with SABV strain SPH114202, showing identity throughout the genome and variant sites (black lines). B) Maximum-likelihood tree of SP2019-01 (red box) based on the alignment of arenavirus sequences. Tree was rooted in the Haartman Institute virus isolate sequence, and bootstrap values are shown next to the branches. CDS, coding sequence; GC, content of guanosine and cytosine; L, large; S, small; SABV, Sabiá virus. Scale bar indicates nucleotide substitutions per site.
1These authors contributed equally to this article.
Page created: May 19, 2020
Page updated: May 19, 2020
Page reviewed: May 19, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.