Volume 26, Number 8—August 2020
Research
Presence of Segmented Flavivirus Infections in North America
Figure 1
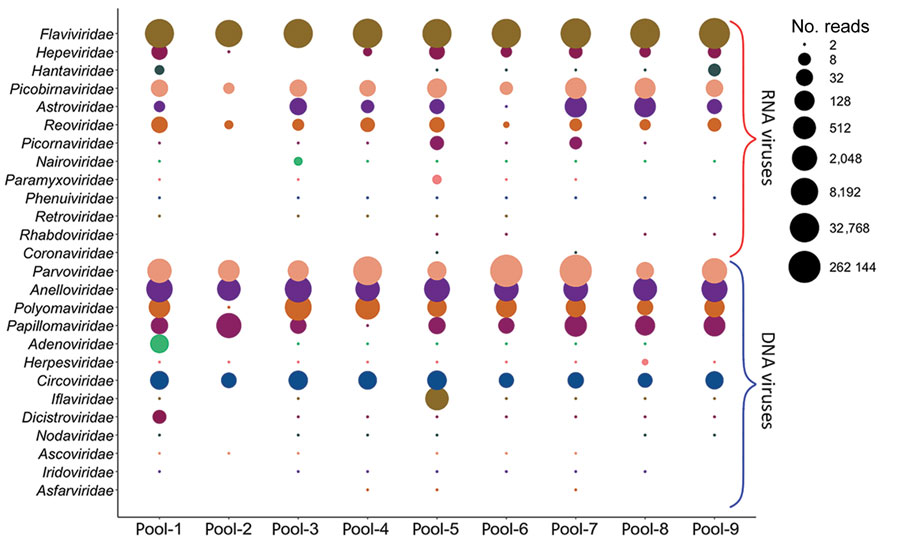
Figure 1. Bubble plot showing the abundance of different viruses in the serum virome of the white-footed mouse. Sequence reads showing the highest sequence similarity to known viruses were normalized as reads per million and were grouped into RNA and DNA virus families. Read numbers were transformed to log2, where the cutoff is ≥2 reads, represented by the smallest circle.
1These authors contributed equally to this article.
Page created: May 14, 2020
Page updated: July 21, 2020
Page reviewed: July 21, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.