Volume 26, Number 8—August 2020
Dispatch
Evolution and Antigenic Drift of Influenza A (H7N9) Viruses, China, 2017–2019
Figure 2
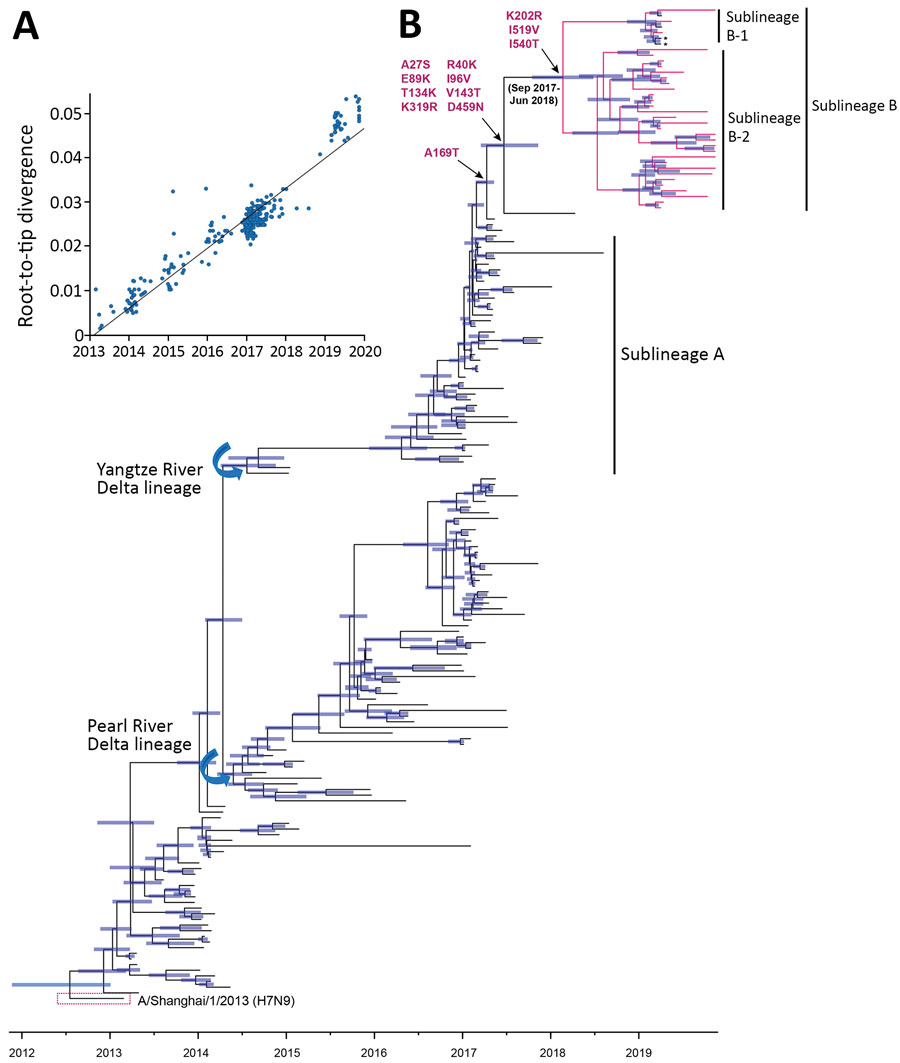
Figure 2. Time-scaled evolution of influenza A(H7N9) viruses, China. A) Analysis of root-to-tip divergence against sampling date for the hemagglutinin gene segment (n = 189). B) Maximum clade credibility tree of the hemagglutinin sequence of H7N9 viruses sampled in China (n = 189); the H7N9 viruses collected in this study are highlighted in red. Asterisk indicates viruses from a human with H7N9 infection within sublineage B during March 2019. Shaded bars represent the 95% highest probability distribution for the age of each node. Parallel amino acid changes along the trunk are indicated.
1These authors contributed equally to this article.
Page created: June 12, 2020
Page updated: July 19, 2020
Page reviewed: July 19, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.