CrAssphage as a Novel Tool to Detect Human Fecal Contamination on Environmental Surfaces and Hands
Geun Woo Park

, Terry Fei Fan Ng, Amy L. Freeland, Vincent C. Marconi, Julie A. Boom, Mary A. Staat, Anna Maria Montmayeur, Hannah Browne, Jothikumar Narayanan, Daniel C. Payne, Cristina V. Cardemil, Aimee Treffiletti, and Jan Vinjé
Author affiliations: Centers for Disease Control and Prevention, Atlanta, Georgia, USA (G.W. Park, T.F.F. Ng, A.L. Freeland, J. Narayanan, D.C. Payne, C.V. Cardemil, A. Treffiletti, J. Vinjé); Atlanta Veteran Administration Medical Center, Atlanta (V.C. Marconi); Emory University School of Medicine, Atlanta (V.C. Marconi); Texas Children’s Hospital, Houston, Texas, USA (J.A. Boom); Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA (M.A. Staat); Cherokee Nation Assurance, Arlington, Virginia, USA (A.M. Montmayeur); Oak Ridge Institute for Science and Education, Oak Ridge, Tennessee, USA (H. Browne)
Main Article
Figure 2
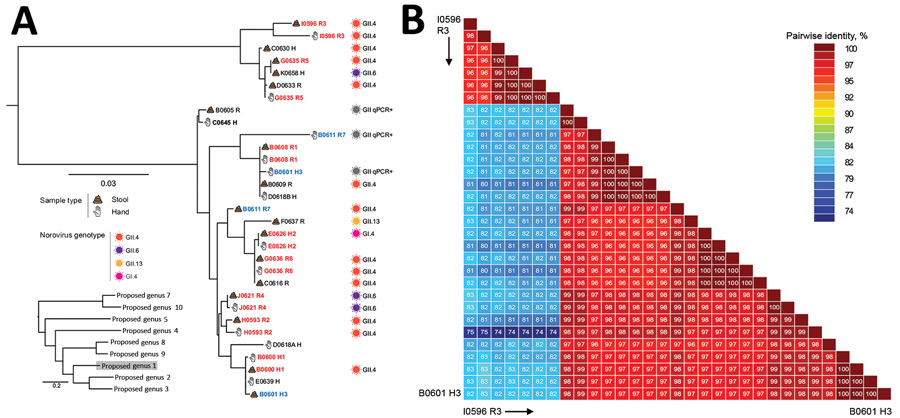
Figure 2. Phylogenetic relationships and pairwise sequence comparison of crAssphage strains from hand rinse samples collected during norovirus outbreaks at long-term care facilities. A) Phylogeny of crAssphage strains. Strain identification includes facility (A–J), strain number, and human source (H, healthcare worker; R, resident) for each isolate. Sample source and genotypes are indicated. Red strain names indicate that both hand and stool sample are genetically related, blue strain names that paired hand and stool samples are genetically distinct. Black strain names indicate hand or stool sample pairs that tested negative for crAssphage. Inset shows position of long-term care strains among reference strains; scale bar indicates number of nucleotide changes between sequences. (B) Color-coded pairwise identity matrix for crAssphage strains. Each cell includes the percentage identity among 2 sequences (horizontally to the left and vertically at the bottom). Key indicates pairwise identity percentages
Main Article
Page created: May 07, 2020
Page updated: July 17, 2020
Page reviewed: July 17, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
