Volume 27, Number 1—January 2021
Dispatch
Seventh Pandemic Vibrio cholerae O1 Sublineages, Central African Republic
Cite This Article
Citation for Media
Abstract
Four cholera outbreaks were reported in the Central African Republic during 1997–2016. We show that the outbreak isolates were Vibrio cholerae O1 serotype Inaba from 3 seventh pandemic El Tor sublineages originating from West Africa (sublineages T7 and T9) or the African Great Lakes Region (T10).
Cholera is a life-threatening diarrheal disease caused by the bacterium Vibrio cholerae, which produces cholera toxin. The seventh cholera pandemic, caused by V. cholerae O1 biotype El Tor, began in Indonesia in 1961 and reached Africa in 1970 (1). Fifty years later, >100,000 cases of cholera are reported annually in sub-Saharan Africa (2).
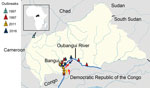
The Central African Republic (CAR) is a landlocked and resource-limited country in central Africa; it was ranked 188/189 on the United Nations Human Development Index in 2018 (http://hdr.undp.org). CAR is relatively large but has a low population density; 2019 data estimate ≈4.75 million inhabitants, or 7.75 persons/km2 (Macrotrends LLC, https://www.macrotrends.net). CAR largely has been spared by the cholera epidemic; only 4 outbreaks had been reported by 2020 (Table 1; Figure 1). The first 2 cholera outbreaks occurred during the same month in 1997 (4). In the first, 443 cases and 88 deaths were reported in southern CAR, along the Oubangui River, close to the border with the Democratic Republic of the Congo (DRC). In the second, 113 cases and 19 deaths were reported in northern CAR, close to the borders with Cameroon and Chad, after which cholera cases continued to be detected in southern CAR, along the Oubangui River. According to reports from the International Federation of Red Cross and Red Crescent Societies, 172 cholera cases and 16 deaths were reported in the region in 2011 (https://reliefweb.int/sites/reliefweb.int/files/resources/MDRCF009finrep.pdf) and 265 cases and 20 deaths were reported in 2016 (https://reliefweb.int/report/central-african-republic/central-africa-republic-cholera-epidemic-outbreak-dref-operation).
The Institut Pasteur de Bangui in CAR performed microbial analyses to confirm the causal agent of these outbreaks and identified 30 V. cholerae O1 serotype Inaba isolates collected during 1997–2016 (Appendix 1 Table 1). We used whole-genome sequencing to fully characterize all 30 V. cholerae O1 isolates in terms of virulence and antimicrobial resistance determinants. We also placed these genomes within a broader phylogenetic context to elucidate their origins and evolutionary history.
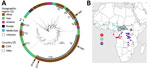
The 30 V. cholerae O1 isolates were received at the Institut Pasteur, Paris, France. We performed antimicrobial susceptibility testing, whole-genome sequencing, comparative genomics, and phylogenetic analyses by using methods previously described (3,5–11) (Appendix 2). We then contextualized these 30 V. cholerae O1 isolates within a global collection of 1,185 seventh pandemic V. cholerae El Tor (7PET) genomic sequences and constructed a maximum-likelihood phylogeny of 1,215 genomes by using 9,964 single-nucleotide variants (SNVs) evenly distributed over the nonrepetitive, nonrecombinant core genome (Figure 2, panel A). Our phylogenomic analysis (Appendix 1 Tables 2–4) showed that all CAR isolates belonged to the 7PET lineage (12,13).
Previous genomic studies described 12 introductions of the 7PET lineage from Southern Asia into Africa during 1970–2016 (3,6). The introduced sublineages were called T1 and T3–T13. The 2 cholera outbreaks in CAR in 1997 were caused by sublineage T7, which had been introduced into West Africa during the early 1980s, and T9, which was introduced in the late 1980s, according to Weill et al. (3). T9 isolates were identified in neighboring countries such as Chad and Cameroon (particularly northern Cameroon) before they were detected in northwest CAR, but T7 isolates were identified in Gabon and the western part of DRC, along the Congo River in Kinshasa, before being identified in CAR along the Oubangui River, a tributary of the Congo River (Figure 2, panel B; Appendix 2 Figures 1, 2).
The 2011 and 2016 outbreaks were caused by closely related bacterial populations from the same sublineage, T10, introduced into East Africa during the 1990s and later detected in the African Great Lakes Region (AGLR) (3) (Figure 2, panel B; Appendix 2 Figure 3). The prevalent T10 sublineage has several clades, and the 2011 and 2016 CAR isolates are characterized by multilocus sequence type (ST) 515, a single-locus variant of ST69; ST69 is the predominant ST among 7PET isolates. In addition, these isolates have an alteration to the wbeT gene, a 4-nucleotide deletion called B01 (Tables 1,2), that underly the Inaba serotype. Phylogenetic data showed that the 7PET strains causing the 2011 and 2016 cholera outbreaks in CAR spread from AGLR to the western part of the DRC and CAR (Figure 2, panel B; Appendix 2 Figure 3). These genomic data are consistent with results from a recent epidemiologic study showing the spatiotemporal distribution of cholera cases during the 2011–2012 and 2015–2017 outbreaks in DRC, suggesting a spread from cholera hotspots in AGLR to major cities in the upstream section of the Congo River, followed by a downstream spread toward the densely populated CAR capital, Kinshasa, and then to the mouth of the Congo River, which opens into the Gulf of Guinea (14).
Except during the outbreak in northwest CAR in 1997, all cholera cases were reported along or close to the Oubangui River, suggesting that 7PET strains probably moved from area to area along the river and with the displacement of human populations. The risk factors in these remote areas are unknown, but the prevailing conditions, such as poor hygiene and sanitary conditions, overcrowding, lack of latrines, and drinking water from the Oubangui River, likely would increase the risk for transmission via the fecal–oral route, as evidenced by the high attack rates observed at several sites when the 2011 outbreak began (15). Nevertheless, since the declaration of the first case in 1997, the small number (<1,000) of cholera cases in CAR contrasts with the much larger numbers in central Africa (2). The low population density of CAR, its poor transport infrastructure, and poor trading links are probably key factors limiting disease spread (15). Phylogenetic analyses showed no other isolates from Africa were derived from CAR isolates in the aftermath of the 4 outbreaks, which also suggests that the transmission of cholera is impeded in this country. Of note, all 4 outbreaks were caused by serotype Inaba 7PET strains. This serotype has a nonmethylated form of lipopolysaccharide caused by an alteration to the wbeT gene (3) (Table 2). The implication of this serotype in all 4 outbreaks suggests that these 7PET sublineages circulated regionally for some time, long enough to acquire this alteration to the wbeT gene, before reaching CAR (Appendix 2 Figures 1–3).
All CAR isolates in this study displayed resistance to polymyxin B, consistent with the susceptibility pattern reported for the El Tor biotype until recently (6). All but 1 of the isolates collected along the Oubangui River in 1997 were susceptible to all other antimicrobial drugs tested; the outlying isolate contained the extended-spectrum β-lactamase blaSHV-2a gene on an IncA/C2 plasmid (Table 1; Appendix 1 Table 3). No susceptible isolates have been collected in CAR since. All the other isolates display mutations of the VC_0715 and VC_A0637 genes, conferring nitrofuran resistance, and carry an SXT/R391 genomic element called ICEVchInd5, encoding resistance to streptomycin (strAB), sulfonamides (sul2), trimethoprim, the O/129 vibriostatic agent (dfrA1), and trimethoprim-sulfamethoxazole (sul2 and dfrA1). The 2011 and 2016 CAR isolates also recently acquired gyrA mutations (Table 1), resulting in resistance to nalidixic acid (Appendix 1 Table 3).
Strains from 3 7PET sublineages caused 4 cholera outbreaks identified in CAR during 1997–2016. The southern and southeastern parts of CAR are higher risk areas for cholera outbreaks, particularly when cases are reported in the western part of DRC. These findings highlight the need for an effective surveillance system, and for coordinated communication actions on cholera that target healthcare professionals and the populations living along the Oubangui River, to prevent and control cholera outbreaks in CAR.
Dr. Breurec is a medical microbiologist on the faculty of medicine and the University Medical Center of Pointe-à-Pitre, Guadeloupe, and at the Institut Pasteur International Network. His research interests include genomic epidemiology, resistance, and the virulence determinants of bacterial pathogens.
Acknowledgment
This work was supported by the Institut Pasteur, Santé Publique France and the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (grant no. ANR-10-LABX-62-IBEID).
References
- Barua DWBG, editor. Cholera. New York: Plenum; 1992.
- Lessler J, Moore SM, Luquero FJ, McKay HS, Grais R, Henkens M, et al. Mapping the burden of cholera in sub-Saharan Africa and implications for control: an analysis of data across geographical scales. Lancet. 2018;391:1908–15. DOIPubMedGoogle Scholar
- Weill FX, Domman D, Njamkepo E, Tarr C, Rauzier J, Fawal N, et al. Genomic history of the seventh pandemic of cholera in Africa. Science. 2017;358:785–9. DOIPubMedGoogle Scholar
- Germani Y, Quilici ML, Glaziou P, Mattera D, Morvan J, Fournier JM. Emergence of cholera in the Central African Republic. Eur J Clin Microbiol Infect Dis. 1998;17:888–90. DOIPubMedGoogle Scholar
- Abubakar A, Bwire G, Azman AS, Bouhenia M, Deng LL, Wamala JF, et al. Cholera epidemic in south Sudan and Uganda and need for international collaboration in cholera control. Emerg Infect Dis. 2018;24:883–7. DOIPubMedGoogle Scholar
- Weill FX, Domman D, Njamkepo E, Almesbahi AA, Naji M, Nasher SS, et al. Genomic insights into the 2016-2017 cholera epidemic in Yemen. Nature. 2019;565:230–3. DOIPubMedGoogle Scholar
- Irenge LM, Ambroise J, Mitangala PN, Bearzatto B, Kabangwa RKS, Durant JF, et al. Genomic analysis of pathogenic isolates of Vibrio cholerae from eastern Democratic Republic of the Congo (2014-2017). PLoS Negl Trop Dis. 2020;14:
e0007642 . DOIPubMedGoogle Scholar - Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–61. DOIPubMedGoogle Scholar
- Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:
e15 . DOIPubMedGoogle Scholar - Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. DOIPubMedGoogle Scholar
- Domman D, Quilici ML, Dorman MJ, Njamkepo E, Mutreja A, Mather AE, et al. Integrated view of Vibrio cholerae in the Americas. Science. 2017;358:789–93. DOIPubMedGoogle Scholar
- Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–5. DOIPubMedGoogle Scholar
- Ingelbeen B, Hendrickx D, Miwanda B, van der Sande MAB, Mossoko M, Vochten H, et al. Recurrent cholera outbreaks, Democratic Republic of the Congo, 2008–2017. Emerg Infect Dis. 2019;25:856–64. DOIPubMedGoogle Scholar
- Penguele A, Djeintote M, Balekouzou A, Tembeti J, Feilema P, Kazambu D, et al. Cholera outbreak investigation in the Central African Republic, October–November 2011 [cited 2020 Feb 23]. https://www.cdcfoundation.org/sites/default/files/upload/pdf/2011CholeraOutbreakReport.pdf
Figures
Tables
Cite This ArticleOriginal Publication Date: December 17, 2020
1These first authors contributed equally to this article.
2Deceased.
3These authors were joint principal investigators.
Table of Contents – Volume 27, Number 1—January 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
François-Xavier Weill, Unité des Bactéries pathogènes Entériques, Institut Pasteur, 28 rue du docteur Roux, 75724 Paris CEDEX 15, France
Top