Volume 27, Number 11—November 2021
Dispatch
Isolation of Novel Mycobacterium Species from Skin Infection in an Immunocompromised Person
Figure 2
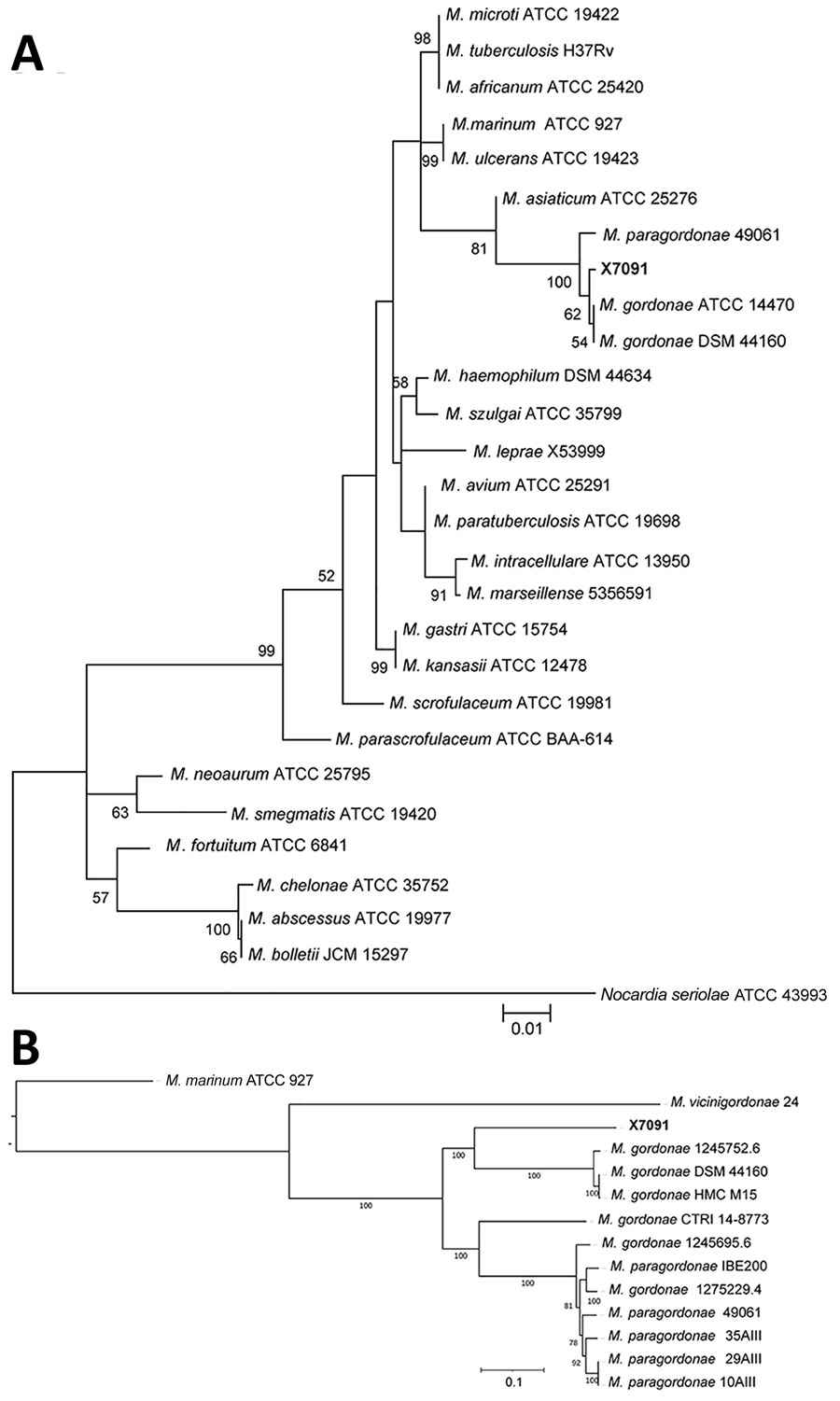
Figure 2. Phylogenetic trees of isolate from novel Mycobacterium gordonae–like infection in a 63-year-old man in China (X7091) and reference isolates. A) Evolutionary tree involving 16S rRNA gene (1,067 positions) of isolate X7091 and 26 Mycobacterium strains. Tree constructed using the maximum-likelihood method and Tamura-Nei model with 500 bootstrap replications in MEGA X (https://www.megasoftware.net). We selected Norcadia seriolae ATCC 43993 as the outgroup. B) Core genome–based maximum-likelihood phylogeny of isolate X7091 and other M. gordonae–like strains analyzed by Roary (https://sanger-pathogens.github.io/Roary) and constructed with a general time-reversible plus gamma maximum model (500 bootstrap replications) using the RaxML tool (14). We selected Mycobacterium marinum ATCC 927 as the outgroup. Scale bars indicate the number of nucleotide substitutions per site.
References
- Itoh S, Kazumi Y, Abe C, Takahashi M. Heterogeneity of RNA polymerase gene (rpoB) sequences of Mycobacterium gordonae clinical isolates identified with a DNA probe kit and by conventional methods. J Clin Microbiol. 2003;41:1656–63. DOIPubMedGoogle Scholar
- Kim BJ, Hong SH, Kook YH, Kim BJ. Mycobacterium paragordonae sp. nov., a slowly growing, scotochromogenic species closely related to Mycobacterium gordonae. Int J Syst Evol Microbiol. 2014;64:39–45. DOIPubMedGoogle Scholar
- Liu G, Yu X, Luo J, Hu Y, Dong L, Jiang G, et al. Mycobacterium vicinigordonae sp. nov., a slow-growing scotochromogenic species isolated from sputum. Int J Syst Evol Microbiol. 2021;71:71. DOIPubMedGoogle Scholar
- Loret JF, Dumoutier N. Non-tuberculous mycobacteria in drinking water systems: A review of prevalence data and control means. Int J Hyg Environ Health. 2019;222:628–34. DOIPubMedGoogle Scholar
- Barber TW, Craven DE, Farber HW. Mycobacterium gordonae: a possible opportunistic respiratory tract pathogen in patients with advanced human immunodeficiency virus, type 1 infection. Chest. 1991;100:716–20. DOIPubMedGoogle Scholar
- Al-Busaidi I, Wong D, Boggild AK. Cutaneous Mycobacterium gordonae infection in an elderly diabetic returned traveller. J Travel Med. 2017;24:24. DOIPubMedGoogle Scholar
- Chen Y, Jiang J, Jiang H, Chen J, Wang X, Liu W, et al. Mycobacterium gordonae in patient with facial ulcers, nosebleeds, and positive T-SPOT.TB test, China. Emerg Infect Dis. 2017;23:1204–6. DOIPubMedGoogle Scholar
- Cheung CY, Cheng NHY, Ting WM, Chak WL. Mycobacterium paragordonae: a rare cause of peritonitis in a peritoneal dialysis patient. Clin Nephrol. 2017;88:371–2. DOIPubMedGoogle Scholar
- Takajo I, Iwao C, Aratake M, Nakayama Y, Yamada A, Takeda N, et al. Pseudo-outbreak of Mycobacterium paragordonae in a hospital: possible role of the aerator/rectifier connected to the faucet of the water supply system. J Hosp Infect. 2020;104:545–51. DOIPubMedGoogle Scholar
- Jagielski T, Borówka P, Bakuła Z, Lach J, Marciniak B, Brzostek A, et al. Genomic insights into the Mycobacterium kansasii complex: an update. Front Microbiol. 2020;10:2918. DOIPubMedGoogle Scholar
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. DOIPubMedGoogle Scholar
- Zong Z. Genome-based taxonomy for bacteria: a recent advance. Trends Microbiol. 2020;28:871–4. DOIPubMedGoogle Scholar
- Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- Luo T, Xu P, Zhang Y, Porter JL, Ghanem M, Liu Q, et al. Population genomics provides insights into the evolution and adaptation to humans of the waterborne pathogen Mycobacterium kansasii. Nat Commun. 2021;12:2491. DOIPubMedGoogle Scholar
1These first authors contributed equally to this article.