Volume 27, Number 12—December 2021
Dispatch
SARS-CoV-2 Variants, South Sudan, January–March 2021
Figure 1
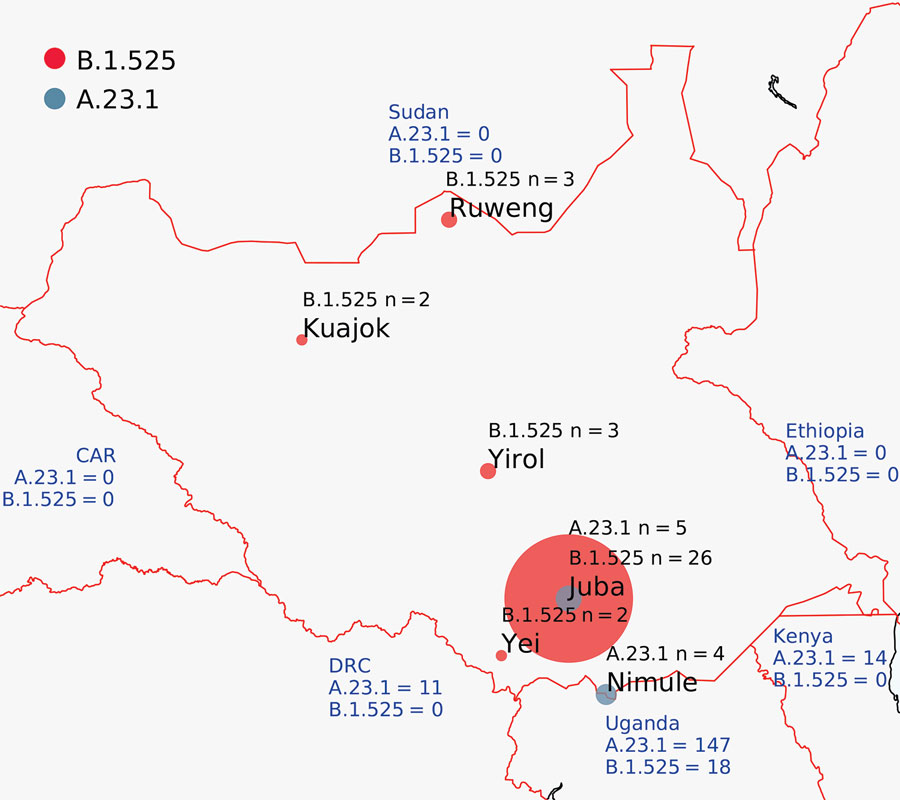
Figure 1. Locations of severe acute respiratory syndrome coronavirus 2 infection case-patients from whom genomes were isolated, South Sudan. Red circles indicate viruses of lineage B.1.525; dark gray circles indicate lineage A.23.1. Circle size is proportional to number of genomes. Blue text shows the number of A.23.1 and B.1.525 genomes reported from neighboring countries. CAR, Central African Republic; DRC, Democratic Republic of the Congo.
Page created: September 11, 2021
Page updated: November 19, 2021
Page reviewed: November 19, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.