Volume 27, Number 4—April 2021
Dispatch
Genomic Characterizations of Clade III Lineage of Candida auris, California, USA
Cite This Article
Citation for Media
Abstract
Candida auris is an emerging multidrug-resistant yeast. We describe an ongoing C. auris outbreak that began in October 2019 in Los Angeles, California, USA. We used genomic analysis to determine that isolates from 5 of 6 patients belonged to clade III; 4 isolates were closely related.
Candida auris was isolated from a patient in Tokyo, Japan in 2009 (1), although clinical isolates have been retrospectively identified from as early as 1996 (2). Since then, bloodstream and other invasive infections caused by C. auris have been reported worldwide (3–5). Many strains of C. auris are multidrug-resistant; some strains require elevated MICs to azoles, echinocandins, and polyenes. In 2019, the US Centers for Disease Control and Prevention (CDC) listed C. auris as an urgent threat to public health (6), highlighting the need for active surveillance and appropriate infection prevention.
Whole-genome sequencing (WGS) and phylogenetic analyses have revealed >4 major clades of C. auris; each clade covers a distinct geographic area, giving C. auris a global distribution (7,8). Researchers have documented several C. auris outbreaks in the United States, mostly caused by strains belonging to clades I and IV (9). We describe several cases of C. auris colonization and infection in patients of long-term acute-care (LTAC) facilities in and around Los Angeles, California, USA.
We screened patients at high risk for drug-resistant infections who were transferred to University of California, Los Angeles (UCLA)–affiliated hospitals from LTAC and skilled nursing facilities (SNFs). We analyzed swab samples of patients’ axilla and groin and yeast isolates from positive fungal culture of clinical specimens using PCR selective for the ITS2 region of the C. auris genome. We conducted antifungal susceptibility testing using broth microdilution; WGS using Illumina MiSeq (Illumina, https://www.illumina.com); and k-mer and single-nucleotide polymorphism (SNP) analyses using CLC Genomics Workbench (QIAGEN, https://www.qiagen.com) and Geneious Prime (Geneious, https://www.geneious.com) (Appendix 1).
During September 2019–September 2020, we screened 113 patients using in-house PCR selective for C. auris according to Los Angeles County Public Health and CDC guidelines (Appendix 1). Six patients tested positive for C. auris with cycle threshold (Ct) values of 22.6–39.7 (Table 1). Patient A tested positive in October 2019; patients B–F tested positive during July–September 2020.
The 6 patients were residents of 4 LTAC facilities in Los Angeles County. All 6 had a history of tracheostomy. Patients A and F had prior history of C. auris colonization; patient F had active infection of a bronchopulmonary fistula. Patient D had C. auris and severe acute respiratory syndrome coronavirus 2 co-infection (Table 1). We cultured C. auris isolates from inguinal and axillary swab samples of patients A, C, D, and E; pleural fluid of patient F; and tracheal aspirate of patient A. The sample from patient A produced few colonies; we treated the patient for bacterial pneumonia. We were not able to isolate C. auris from patient B (Ct = 39.5).
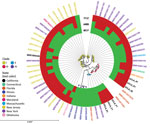
All C. auris isolates were resistant to amphotericin B (MIC = 2 μg/mL) and fluconazole (MIC >64 μg/mL) but susceptible to echinocandins (Table 2). We conducted k-mer analysis using 261 C. auris sequences available on GenBank, most of which were described previously (10) (Appendix 2 Table 1). All 6 UCLA isolates belonged to clade III (Appendix 2 Table 1). We conducted a phylogenetic analysis of clade III isolates using k-mers (Appendix 1 Figure).
In the United States, researchers have identified isolates belonging to all 4 clades; although these isolates show geographic relationships (9), clade I is predominant across the country. Clade III isolates have been identified in Indiana, Texas (11), and Florida. We conducted a k-mer–based phylogenetic analysis of C. auris isolates in the United States (Figure). SNP analysis showed that 5 of the UCLA isolates were closely related (3–12 SNPs); isolate F1 was genetically distinct (77–79 SNPs). All 6 isolates were distinct from isolates from Indiana (65–139 SNPs) and Florida (47–117 SNPs) (Appendix 1 Table 1).
We also analyzed the sequences of 2 genes associated with antifungal resistance: erg11 (lanosterol 14-α demethylase) and fks1 (subunit of 1,3-β-D-glucan synthase). Sequences of erg11 were identical among all isolates, with 99.6% pairwise nucleotide identity to the reference (GenBank accession no. CP043531) and 2 amino acid substitutions: V125A and F126L (Appendix 1 Table 2). Mutations at aa 126 are associated with increased azole resistance in C. auris (7) and are a common feature of clade III isolates (12). The F126L mutation appears to be exclusive to clade III (10). These findings are consistent with results of antifungal susceptibility testing, which showed that all isolates were resistant to fluconazole (Table 2). Sequences of fks1 were identical in 5 isolates (A1, A2, C1, D1, E1), with 99.9% pairwise nucleotide identity to the reference (GenBank accession no. CP043531); these isolates had 1 amino acid substitution: I1572L (Appendix 1 Table 2). Isolate F1 had the same substitution in addition to I1095L. All isolates had a wild-type serine at aa 639; mutations at this location are linked to echinocandin resistance in C. auris (13). All isolates were susceptible to caspofungin, micafungin, and anidulafungin.
To identify and prevent the spread of C. auris in this hospital system, we used an in-house PCR to screen patients for this pathogen. WGS of isolates from patients transferred from LTAC facilities revealed that these isolates are closely related, suggesting an ongoing outbreak with community spread in the Los Angeles area.
The isolates described here were all resistant to fluconazole and amphotericin B but susceptible to echinocandins according to the CDC tentative breakpoints (https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html). In addition, all isolates had an F126L mutation in the erg11 gene, which is unique to clade III strains and associated with fluconazole resistance (10).
Patient D was admitted to an SNF after complications from pneumonia caused by coronavirus disease (COVID-19). Few cases of C. auris and COVID-19 co-infection have been reported (14,15). After COVID-19 infection, patient D had multiple complications requiring a tracheostomy and enteral feeding tube; the patient was subsequently transferred to an LTAC for rehabilitation. A substantial portion of adult patients who recover from severe COVID-19 have long-term sequelae and might require admission to SNFs or LTACs. Therefore, the COVID-19 pandemic might lead to increased transmission of C. auris in SNFs because of increased admissions and shortages of personal protective equipment. During critical shortages, CDC guidelines permit extended use of isolation gowns for patients who are known to be infected with the same infectious disease if there are no additional known coinfections transmitted through contact (https://www.cdc.gov/coronavirus/2019-ncov/hcp/non-us-settings/emergency-considerations-ppe.html#ppe-specific-strategies). To encourage appropriate use of personal protective equipment and prevent transmission, it is essential that facilities screen patients for C. auris.
One limitation of this study is the lack of additional epidemiologic history of the patients, especially in the context of travel-related exposures. The ability to track cases to a location with known outbreaks of clade III C. auris strains is essential to determining the origin of the current outbreak. Further investigation is needed to explain why patient F had a genetically distinct isolate, suggesting a separate introduction.
In conclusion, we identified a unique clade III C. auris strain in an ongoing outbreak in LTAC facilities since 2019. These findings indicate active community spread of multidrug-resistant C. auris in the Los Angeles area.
Dr. Price is a clinical microbiology fellow at the David Geffen School of Medicine at University of California, Los Angeles in Los Angeles, California, USA. His research interests include microbial genomics, fungal species identification, and antimicrobial resistance mechanisms.
References
- Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53:41–4. DOIPubMedGoogle Scholar
- Forsberg K, Woodworth K, Walters M, Berkow EL, Jackson B, Chiller T, et al. Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med Mycol. 2019;57:1–12. DOIPubMedGoogle Scholar
- Calvo B, Melo AS, Perozo-Mena A, Hernandez M, Francisco EC, Hagen F, et al. First report of Candida auris in America: Clinical and microbiological aspects of 18 episodes of candidemia. J Infect. 2016;73:369–74. DOIPubMedGoogle Scholar
- Armstrong PA, Rivera SM, Escandon P, Caceres DH, Chow N, Stuckey MJ, et al. Hospital-associated multicenter outbreak of emerging fungus Candida auris, Colombia, 2016. Emerg Infect Dis. 2019;25:1339–46. DOIPubMedGoogle Scholar
- Rhodes J, Abdolrasouli A, Farrer RA, Cuomo CA, Aanensen DM, Armstrong-James D, et al. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. [Erratum in: Emerg Microbes Infect. 2018;7:104]. Emerg Microbes Infect. 2018;7:43. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. 2019 [cited 2019 Nov 1]. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf
- Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64:134–40. DOIPubMedGoogle Scholar
- Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019;25:1780–1. DOIPubMedGoogle Scholar
- Chow NA, Gade L, Tsay SV, Forsberg K, Greenko JA, Southwick KL, et al.; US Candida auris Investigation Team. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis. 2018;18:1377–84. DOIPubMedGoogle Scholar
- Chow NA, Muñoz JF, Gade L, Berkow EL, Li X, Welsh RM, et al. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. MBio. 2020;11:e03364–19. DOIPubMedGoogle Scholar
- Long SW, Olsen RJ, Nguyen HAT, Ojeda Saavedra M, Musser JM. Draft genome sequence of Candida auris strain LOM, a human clinical isolate from greater metropolitan Houston, Texas. Microbiol Resour Announc. 2019;8:e00532–19. DOIPubMedGoogle Scholar
- Healey KR, Kordalewska M, Jiménez Ortigosa C, Singh A, Berrío I, Chowdhary A, et al. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother. 2018;62:e01427–18. DOIPubMedGoogle Scholar
- Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother. 2018;73:891–9. DOIPubMedGoogle Scholar
- Chowdhary A, Tarai B, Singh A, Sharma A. Multidrug-resistant Candida auris infections in critically ill coronavirus disease patients, India, April–July 2020. Emerg Infect Dis. 2020;26:2694–6. DOIPubMedGoogle Scholar
- Rodriguez JY, Le Pape P, Lopez O, Esquea K, Labiosa AL, Alvarez-Moreno C. Candida auris: a latent threat to critically ill patients with COVID-19. Clin Infect Dis. 2020 Oct 18 [Epub ahead of print].
Figure
Tables
Cite This ArticleOriginal Publication Date: March 11, 2021
Table of Contents – Volume 27, Number 4—April 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Address for correspondence: Shangxin Yang, UCLA Clinical Microbiology Laboratory, 11633 San Vicente Blvd, Los Angeles, CA 90049, USA
Top