Monitoring SARS-CoV-2 Circulation and Diversity through Community Wastewater Sequencing, the Netherlands and Belgium
Ray Izquierdo-Lara, Goffe Elsinga, Leo Heijnen, Bas B. Oude Munnink, Claudia M.E. Schapendonk, David Nieuwenhuijse, Matthijs Kon, Lu Lu, Frank M. Aarestrup, Samantha Lycett, Gertjan Medema
1, Marion P.G. Koopmans
1, and Miranda de Graaf
1
Author affiliations: Erasmus University Medical Center, Rotterdam, the Netherlands (R. Izquierdo-Lara, B.B. Oude Munnink, C.M.E. Schapendonk, D. Nieuwenhuijse, M. Kon, M.P.G. Koopmans, M. de Graaf); KWR Water Research Institute, Nieuwegein, the Netherlands (G. Elsinga, L. Heijnen, G. Medema); University of Edinburgh, Edinburgh, Scotland, UK (L. Lu, S. Lycett); Technical University of Denmark, Kongens Lyngby, Denmark (F.M. Aarestrup)
Main Article
Figure 3
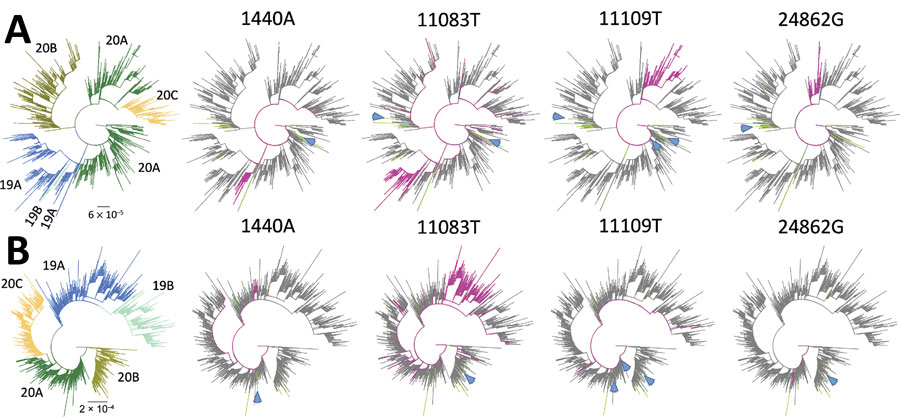
Figure 3. Phylogenetic trees showing 4 low-frequency variants detected in sewage samples in study of severe acute respiratory syndrome coronavirus 2 circulation and diversity through community wastewater sequencing, the Netherlands and Belgium. A) The Netherlands–Belgium subsample; B) global subsample. Patient sequences containing the mutation are shown in magenta. Lines in green indicate sewage samples sequenced in this study. Clades (19A, 19B, 20A, 20B, and 20C) are indicated in colors at the left of the figure. Blue arrows show the consensus sequences (if available) of the sewage samples in which the low-frequency variant was detected. Scale bars indicate the inferred number of nucleotide substitutions per site.
Main Article
Page created: March 02, 2021
Page updated: April 20, 2021
Page reviewed: April 20, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
