Volume 27, Number 6—June 2021
Dispatch
Cutaneous Leishmaniasis Caused by an Unknown Leishmania Strain, Arizona, USA
Figure 2
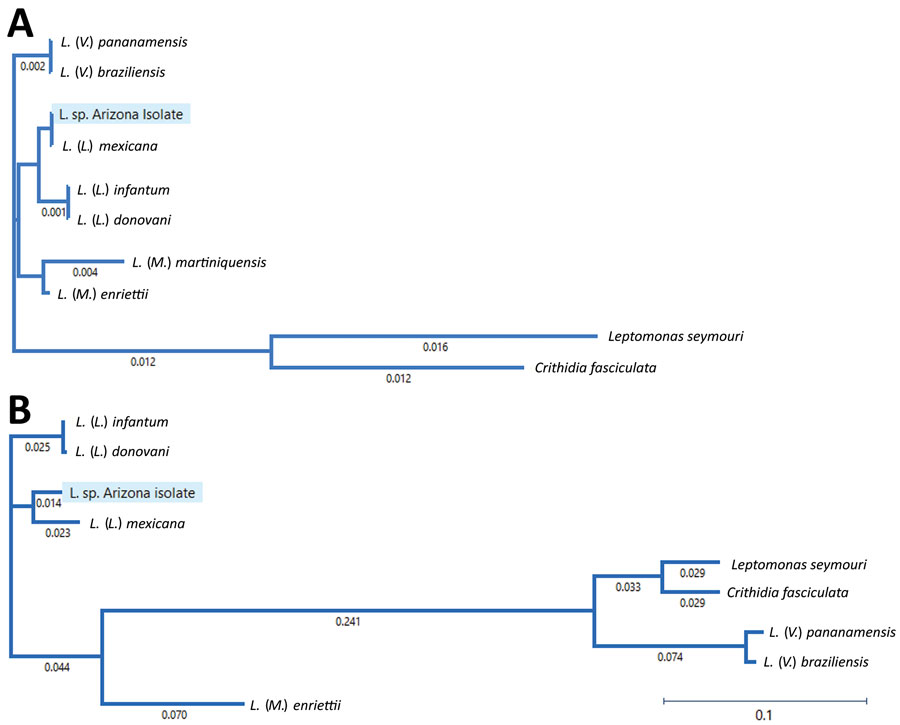
Figure 2. Phylogenetic tree of Leishmania subgenus isolates from a patient in Arizona, USA, and reference Leishmania species in relationship to species in the subgenera Leishmania, Viannia, and Mundina. A) Phylogenetic tree of Leishmania 18S rRNA genes. Sequences of Crithidia fasciculata and Leptomonas seymouri are included as references. L. (V.) panamensis (GenBank accession no. GQ332362); L. (V.) braziliensis (accession no. GQ332355); L. (L) mexinana (accession no. GQ332260); L. (L.) infantum (accession no. GQ332359); L. (L.) donovani (accession no. GQ332356); L. (M.) martiniquensis (accession no. AF303938); L. (M.) enriettii (accession no. ATAF02000704); Leptomonas seymore (accession no. KP717894); and Crithidia fasciculata (accession no. Y00055). The 2 non-Leishmania trypanosomatids (Leptomonas seymore and Crithidia fasciculata) were included in the phylogenetic tree because they were previously described as co-infecting parasites in human leishmaniasis cases. B) Phylogenetic tree of glyceraldehyde-3-phosphate dehydrogenase genes. Sequences from Crithidia fasciculata and Leptomonas seymouri were included as references. Numbers along branches indicate bootstrap values. Scale bars indicate nucleotide substitutions per site.
1Current affiliation: University of Arkansas for Medical Sciences, Little Rock, Arkansas, USA.