Volume 27, Number 7—July 2021
Research
Novel Morbillivirus as Putative Cause of Fetal Death and Encephalitis among Swine
Cite This Article
Citation for Media
Abstract
Morbilliviruses are highly contagious pathogens. The Morbillivirus genus includes measles virus, canine distemper virus (CDV), phocine distemper virus (PDV), peste des petits ruminants virus, rinderpest virus, and feline morbillivirus. We detected a novel porcine morbillivirus (PoMV) as a putative cause of fetal death, encephalitis, and placentitis among swine by using histopathology, metagenomic sequencing, and in situ hybridization. Phylogenetic analyses showed PoMV is most closely related to CDV (62.9% nt identities) and PDV (62.8% nt identities). We observed intranuclear inclusions in neurons and glial cells of swine fetuses with encephalitis. Cellular tropism is similar to other morbilliviruses, and PoMV viral RNA was detected in neurons, respiratory epithelium, and lymphocytes. This study provides fundamental knowledge concerning the pathology, genome composition, transmission, and cellular tropism of a novel pathogen within the genus Morbillivirus and opens the door to a new, applicable disease model to drive research forward.
Paramyxoviridae encompasses a group of large (300–500 nm in diameter), enveloped, pleomorphic viruses with RNA genomes of 14.6–20.1 kb. The family comprises of 4 subfamilies and 17 genera that contain >70 species and includes global human and animal viral pathogens of concern (1). Currently, the genus Morbillivirus, in subfamily Orthomyxovirinae, contains measles virus (MeV), rinderpest virus (RPV), peste des petits ruminants virus (PPRV), canine distemper virus (CDV), phocine distemper virus (PDV), cetacean morbillivirus (CMV), and feline morbillivirus (FeMV) (2,3).
Morbillivirus genomes encode 6 structural proteins in the following order: nucleocapsid (N) protein, phosphoprotein (P), matrix (M) protein, hemagglutinin (H) protein, fusion (F) protein, and large polymerase (L) protein (2). Two nonstructural proteins, C and V, are expressed from the P open reading frame and are thought to interfere with the innate immune response in at least a subset of members of the family Paramyxoviridae (4).
Morbilliviruses cause respiratory and gastrointestinal disease and profound immune suppression (5). Morbillivirus host species experience a similar pathogenesis; infection occurs through inhalation, direct contact with body fluids, or fomites or vertical transmission (6–8). Carnivore morbilliviruses readily invade the central nervous system (CNS), and all morbilliviruses produce intranuclear viral inclusion bodies containing nucleocapsid-like structures (1,9,10).
Paramyxoviruses known to naturally infect swine include porcine rubulavirus, Menangle virus, Nipah virus, and porcine parainfluenza virus (11–16). Less well-characterized paramyxoviruses associated with central nervous and respiratory disease in pigs also have been reported (17–20), but none of these viruses are classified in the genus Morbillivirus. Using histopathology, metagenomic sequencing, and RNA in situ hybridization (ISH), we identified a novel morbillivirus in swine as the putative cause of an outbreak of reproductive disease characterized by fetal mummification, encephalitis, and placentitis.
Clinical Background and Samples
In early 2020, the Iowa State University Veterinary Diagnostic Laboratory (Ames, IA, USA) received 22 porcine fetuses from 6 litters (A–F) that originated from a commercial breeding herd in northern Mexico for routine diagnostic investigation (Table 1). The breeding herd was comprised 2,000 sows and reported reproductive clinical signs characterized by an increased percentage (18% reported) of mummified fetuses and stillbirths. For negative controls, we used fetal tissues from 2 litters from a 3,000-head sow farm in the United States that was experiencing increased mummified fetuses and stillborn fetuses.
Pathology
At necropsy, we recorded the condition, including neonatal morality, mummified fetus, moderate autolysis, or stillbirth, and the crown-to-rump length (CRL) of each fetus or piglet in the case record (Appendix). We processed fixed tissues by standard technique, stained tissues by using hematoxylin and eosin, and performed histologic evaluations. We used sections from the paraffin blocks for ISH.
Porcine Morbillivirus ISH and PCR
At Iowa State University Veterinary Diagnostic Laboratory, we used RNAscope 2.5 HD Reagent Kit (Advanced Cell Diagnostics [ACD] bio-techne, https://acdbio.com) to perform RNA ISH according to the manufacturer’s instructions for formalin-fixed paraffin-embedded samples (Appendix). We prepared fetal thoracic tissue homogenate from each litter, extracted nucleic acids, and performed PCR similar to previously described methods (21). We used fetal heart and lung to perform PCRs for porcine circovirus 2 and 3 (PCV2 and PCV3), porcine parovirus 1 (PPV1), and porcine reproductive and respiratory virus (PRRSV). We used kidney tissue for Leptospira sp. PCR (Appendix).
We developed a real-time reverse transcription PCR (RT-PCR) specific for PoMV by using the MBLV-900F and MBLV-988R primers and the MBL-959P probe (Appendix Table 2). In addition, we performed a previously described real-time RT-PCR (22) to rule out porcine rubulavirus coinfection (Appendix).
Metagenomics and Bioinformatics Analysis
We extracted total nucleic acid of 2 pooled fetal thoracic tissue samples and prepared sequencing libraries, as described previously (23). The first pool consisted of litters A and B; the second pool consisted of litters D and E. We used the MiSeq platform (Illumina, https://www.illumina.com) to sequence the libraries by using the MiSeq 600-Cycle Reagent Kit v3 (Illumina). We preprocessed raw sequencing reads and classified reads by using Kraken version 0.10.5-β (24) with the standard database. We used Kaiju version 1.6.2 (25) to classify unclassified reads, and used KronaTools-2.6 (26) to generate the interactive html charts for hierarchical classification results. We extracted reads of the virus of interest, morbillivirus, from the classification results for de novo assembly by using ABySS version 1.3.9 (27), iva version 1.0.8 (28), and Spades version 3.11.1 (29). We manually refined the resulting contigs, and then curated and elongated contigs by using BLAST (https://blast.ncbi.nlm.nih.gov), SeqMan Pro (https://seqman.software), and integrated genomics viewer for visualization (30). We closed the genome gap by conventional RT-PCR with specifically designed primers.
We used ClustalW (http://www.clustal.org) to generate multiplex sequence alignments. We constructed phylogenetic trees based on whole-genome sequences and amino acid sequences of the L protein from aligned sequences by the maximum likelihood model in MEGA version X (https://www.megasoftware.net). We used L protein sequences because paramyxoviruses currently are classified based on the sequence comparison of L protein, the RNA-dependent RNA polymerase. We evaluated the robustness of the phylogenetic tree by bootstrapping using 500 replicates. We used interactive Tree of Life (iTOL, https://itol.embl.de) to display, manipulate, and annotate bases of the whole-genome sequence and L protein amino acid sequence trees (31).
Gross Pathology and Pathogen Detection
Among 22 porcine fetuses from the 6 litters (A–F) submitted for diagnostic investigation, CRL length varied from 7 to 29.5 cm (Table 1). We noted the litter identification, sow parity, CRL by individual fetus and piglet, and total number born and number of affected fetuses in each litter as reported by the sow farm (Table 1). Submitted fetuses were 1 neonatal death, in which necropsy revealed aerated lungs; 3 stillbirths, all of which were full-term, fresh-type fetuses but had fetal atelectasis; 14 mummified fetuses, in which in utero death occurred with sufficient time for complete dehydration of tissue; and 4 fetuses with moderate autolysis, in which in utero death occurred without sufficient time for complete dehydration of tissue. Gross evaluation of stillbirths and the single neonatal death was unremarkable.
To address differential diagnoses, we used quantitative PCR (qPCR) to detect known swine viral and bacterial reproductive pathogens. We did not detect PCV2, PCV3, PRRSV, PPV1, or Leptospira sp. by qPCR or quantitative RT-PCR (qRT-PCR) in any litter.
Metagenomic Sequencing
We pooled samples of fetal thoracic tissue from litters A and B (A–B), both of which had encephalitis noted histologically. We also pooled fetal thoracic tissue from litters D and E (D–E), which had leukocytes in the epicardium noted histologically. We performed NGS on the 2 pooled samples by using the MiSeq platform (Illumina). After using an in-house bioinformatics analysis pipeline, we detected and identified 693 paramyxovirus-like reads in A–B and 118,772 in D–E. No reads of other pathogens were identified. De novo assembly obtained 2 contigs with 4,869 and 10,456 nt from pooled sample A–B and another 2 contigs from pooled sample D–E. The nucleotide sequences of the contigs from A–B and D–E were 100% identical but the contigs assembled from pooled sample D–E were slightly longer, 5,042 and 10,705 nt. Sequence analysis of the 4 contigs suggested the presence of a previously undescribed paramyxovirus of genus Morbillivirus. The 2 shortest contigs had <40% nt identity to PDV (GenBank accession no. KC802221) and CDV (GenBank accession no. AF014953) at the 3′ end. The 2 longer contigs had >60% nt identity to PDV and CDV at the 5′ end. We propose this paramyxovirus be named porcine morbillivirus (PoMV).
Genome Sequence Characterization
A complete genome sequence of PoMV (GenBank accession no. MT511667) was obtained by using RT-PCR to fill the gap between the 2 contigs. We designed 14 pairs of primers according to the obtained genome sequence and sequenced the RT-PCR products again, confirming the accuracy of the whole-genome sequence. The genome size of PoMV is 15,714 bases and has a G+C content of 45.19%. The 3′ leader sequence of the PoMV is 55 nt with 13/20 initial nt being highly conserved among morbilliviruses (Appendix Figure 1). PoMV has a 5′ trailer sequence of 41 nt, similar to other morbilliviruses that have a trailer sequence of 40 or 41 nt (Appendix Figure 1), except for FeMV, which has an unusually long trailer sequence of 400 nt. The last 11 nt of 5′ trailer sequences are conserved in all morbilliviruses.
The genome of PoMV contains 6 genes, 3′-N-P/V/C-M-F-H-L-5′, similar to other morbilliviruses. The pairwise alignment of the predicted gene and gene products in PoMV and other paramyxoviruses showed the highest nucleotide and amino acid identities with members of the genus Morbillivirus (Table 2). Nucleotide identities were 56.3%–66.6% for N, 44.3%–61.4% for P, 62.4%–68.4% for M, 50.2%–64.1% for F, 31.7%–52.7% for H, and 58.1%–68.1% for L; amino acid identities were 56.3%–70.3% for N, 26%–50.1% for P, 60.9%–78.8% for M, 42.2%–65.5% for F, 15.7%–45.4% for H, and 56.2%–75.8% for L. PoMV had the highest identities to PDV and CDV and the lowest to FeMV (Table 2).
We noted PoMV included the conserved N terminal motif MA(T/S)L in morbilliviruses containing the sequence MASL in the nucleoprotein (N) (Appendix Figure 2). We identified a leucine-rich motif at aa positions 4–11 and 70–77 in the N protein of PoMV (Appendix Figure 2). We identified 2 initiation codons in the P/V/C gene of PoMV; the first translates P and V and the second translates C. In addition, we identified a UC-rich editing site, ttaaaagggg, in the P/V/C gene of PoMV. We detected a conserved cleavage site, RRQKRF, ≈114 aa residues from the N terminus of the F protein. The F protein of PoMV also contains 9/10 Cys residues and 3 potential N-glycosylation sites.
Phylogenetic Analyses
We constructed phylogenetic trees by using the whole-genome sequences (Figure 1, panel A) and the predicted aa sequences of the L gene, the RNA-dependent RNA polymerase gene of PoMV and other members of Paramyxoviridae (Figure 1, panel B). In both phylogenetic trees, PoMV clustered with other morbilliviruses, with high bootstrap supporting a distinct subgroup (Figure 1). Both phylogenetic analyses also confirmed the findings from the results of pairwise alignment and demonstrated that PoMV was most closely related to CDV and PDV; closely related to CMV, PPRV, MeV, and RPV; and most distantly related to FeMV in the genus Morbillivirus (Figure 1). Overall, these data further support that PoMV is a previously undescribed member in the genus Morbillivirus, subfamily Orthoparamyxovirinae, and family Paramyxoviridae.
Histopathology and PoMV ISH
We performed histologic examination and RNA ISH by litter (Table 3). For all 6 litters, the positive control probe, Ss-PPIB, was positive and the negative control probe, DapB, was negative on the single slide assayed (data not shown). PoMV RNA was not detected by ISH in the cerebrum and cerebellum of 2 stillborn fetuses from unaffected litters used as negative controls (data not shown).
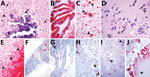
The single stillborn fetus submitted from litter A had multifocal areas of mineralization associated with neuronal necrosis and rarefaction in the cerebrum and brainstem (Figure 2, panel A). Cerebral vessels were occasionally surrounded by lymphocytes. Eosinophilic intranuclear and intracytoplasmic viral inclusion bodies were in neurons (Figure 2, panel A) and glial cells in the cerebrum and internal granular layer of the cerebellum. Rarely, respiratory epithelium lining bronchi and bronchioles contained intranuclear viral inclusion bodies. Histologic evaluation of the heart, spleen, and kidney was diagnostically unremarkable. Moderate autolysis of the liver precluded a thorough histologic evaluation. ISH detected extensive PoMV RNA in the cerebellum in the external granular layer, molecular layer, internal granular layer, and white matter (Figure 2, panel B) and in neurons and axons in the cerebrum (Figure 2, panel C). We also detected PoMV RNA in clusters of respiratory epithelium lining bronchi and bronchioles, scattered lymphocytes and aggregates of lymphocytes in periarteriolar lymphoid sheaths in the spleen, and aggregates of tubular epithelium in rare tubules within the cortex of the kidney. ISH did not detect PoMV RNA in the heart or liver.
The single full-term piglet submitted from litter B had multifocal mineralization and rare satellitosis in the cerebrum (Figure 2, panel D). Adjacent to the lateral ventricle, marked neuropil rarefaction, mineralization, neuronal necrosis, and leukocyte infiltration were visible. Numerous viral inclusion bodies could be seen in neurons (Figure 2, panel D) and glial cells. Histologic evaluation of the cerebellum, lung, heart, spleen, liver, and kidney was diagnostically unremarkable. PoMV was detected by ISH in the gray matter of the cerebrum with extensive labeling; and in the white matter with less but still abundant labeling (Figure 2, panel E). PoMV also was detected by ISH in low to moderate numbers of respiratory epithelium in multifocal bronchi and bronchioles, and individual lymphocytes were noted in the spleen. PoMV RNA was not detected in the heart, kidney, or liver.
Litter C also was represented by a single stillbirth in which histologic evaluation of the cerebrum, cerebellum, lung, heart, and kidney was diagnostically unremarkable. Moderate autolysis of the liver and spleen precluded a thorough histologic evaluation. However, PoMV was detected by ISH in the endothelial cells of a single vessel in the cerebrum, scattered cells within alveolar septa and numerous lymphocytes within the spleen (Figure 2, panel F). PoMV was not detected by ISH in the heart.
Litter D was represented by 3 mummified fetuses and 2 fetuses with moderate autolysis. In a single section of the heart from a fetus with moderate autolysis, we noted mononuclear leukocytes in the epicardium. Lung and kidney of fetuses with moderate autolysis were unremarkable. Autolysis of the spleen and liver from these fetuses and lung, kidney, and heart from mummified fetuses precluded histologic evaluation. Despite severe autolysis and mineralization, leukocytes were observed in the allantoic connective tissue of the placenta. PoMV was detected by ISH in the alveolar septa, bronchi, and bronchioles of the lung of both fetuses with moderate autolysis as well as bronchi and bronchioles of a mummified fetus. PoMV RNA also was detected in scattered lymphocytes in one fetus with moderate autolysis and abundant lymphocytes in the spleen of the other. Mononuclear leukocytes in the allantoic connective tissue and allantoic epithelium of the placenta also contained PoMV RNA (Figure 2, panel G) as did rare renal tubular epithelium and endothelium of a vessel adjacent to the renal pelvis in a mummified fetus (Figure 2, panel H). PoMV was not detected by ISH in the heart or liver of moderately autolyzed fetuses.
Litter E consisted of 9 mummified fetuses and 1 fetus with moderate autolysis. In a single section of 1 heart, the epicardium contained multifocal mononuclear leukocyte aggregates. The lung and kidney were unremarkable in the fetus with moderate autolysis. Autolysis of the heart, lung, and kidney of mummified fetuses precluded histologic evaluation. PoMV was detected by ISH extensively in the conducting airway epithelium and alveolar septa of the lung (Figure 2, panel I), renal tubules, and in rare leukocytes in the epicardium of the fetus with moderate autolysis. ISH was not performed on mummified fetal tissues.
Litter F was represented by a mummified fetus, a fetus with moderate autolysis, and a stillborn fetus. Abundant mononuclear leukocytes were expanding the allantoic connective tissue of the placenta. Histologic evaluation of the cerebrum, cerebellum, spleen, and liver of the stillborn fetus was diagnostically unremarkable. Kidney and lung of the stillborn and moderately autolyzed fetuses were unremarkable. The heart was unremarkable in all fetuses. Autolysis of the mummified fetus precluded evaluation of the lung and kidney. PoMV was detected in the epithelium of conducting airways and alveolar septa of the mummified fetus and the fetus with moderate autolysis. We noted extensive labeling in the allantoic connective tissue and mononuclear leukocytes throughout the placenta (Figure 2, panel J). PoMV was not detected in the lung, heart, or spleen of the stillborn fetus or heart of the fetus with moderate autolysis.
PoMV Real-Time RT-PCR
Fetal thoracic tissues from litters A, B, D, and E were further subjected to real-time RT-PCR to approximate the viral load by litter. All were positive for PoMV with fetal thoracic tissues from litters A and E having a higher viral load. Litter A had a quantification cycle (Cq) value of 19.7; the Cq for litter E was 19.4. Fetal thoracic tissues from litters B and D had a lower viral load values; B had Cq of 23.4 and D had Cq of 20.2. PoMV was not detected by real-time RT-PCR in the fetal thoracic tissues of 2 litters of unaffected fetuses selected as negative controls. In addition, porcine rubulavirus was not detected in any sample by RT-PCR.
We report a novel porcine morbillivirus, PoMV, as a cause of fetal death, encephalitis, and placentitis among 6 swine litters. The synchronous use of 3 independent and complementary lines of evidence, pathology, metagenomic sequencing, and in situ hybridization, aided in PoMV discovery. Although several paramyxoviruses have been found, the existence of a naturally occurring morbillivirus in swine previously was unknown. Analyses of predicted nt and aa sequences of 6 genes revealed that PoMV has the highest nucleotide (31.7%–68.1%) and amino acid (26%–75.2%) identities with members in the genus Morbillivirus in the N, P, M, F, H, and L genes (Table 2). Phylogenetic analyses based on the whole genome sequence (Figure 1, panel A) and amino acid sequence of the L gene (Figure 1, panel B) further demonstrated that PoMV forms a distinct cluster in morbillivirus and is most closely related to PDV and CDV.
Experimental inoculation of CDV and PPRV in domestic pigs has resulted in infection (32,33), but no morbillivirus previously has been known to infect swine naturally. Other viruses within the family Paramyxoviridae that infect swine include porcine rubulavirus (genus Orthorubulavirus, subfamily Rubulavirunae), Menangle virus (genus pararubulavirus, subfamily Rubulavirunae), Nipah virus (genus Henipavirus, subfamily Orthoparamyxovirinae), and porcine parainfluenza virus 1 (genus Respirovirus, subfamily Orthoparamyxovirinae). Among these, only porcine rubulavirus and Menangle virus are thought to cause fetal mummification and stillbirths, as observed with PoMV (34,35). Histologic lesions noted for PoMV are similar to Menangle virus and included encephalitis, viral inclusion bodies, and nonsuppurative myocarditis (36).
Similar to our findings with PoMV, MeV has been reported to be transmitted vertically resulting in premature stillbirth, stillbirth, premature birth, neonatal death, or congenital measles (6,37,38). Viral inclusion bodies have been observed in human congenital MeV infection and MeV was detected in placenta and splenic lymphocytes by immunohistochemistry (39). Herein, viral inclusion bodies were commonly observed in the cerebrum and cerebellum (Table 3; Figure 2, panels A,D) and rarely in the respiratory epithelium lining conducting airways. ISH demonstrated presence of PoMV in the placenta (Figure 2, panels G, J) and splenic lymphocytes (Figure 2, panel F), and in the cerebrum (Figure 2, panels C, E), cerebellum (Figure 2, panel B), lung (Figure 2, panel I), and to a lesser extent in the kidney (Figure 2, panel H) and heart. Of note, the degree of viral involvement within the monochorionic placenta and effects of MeV on human monozygotic twins was inconsistent; 1 in utero death at 32 weeks gestation and 1 surviving infant with no clinical signs of MeV infection (39). This observation, along with the placentation of swine, large litter size, and observations from other swine viral reproductive pathogens (40), likely accounts for the variable effects on litters, which were characterized by fetal and piglet death at various stages of gestation and resulted in fetal mummification, in utero death, and stillbirth, along with the variability of ISH staining observed among fetuses and between litters.
Cellular tropism of PoMV determined by ISH aligns with other morbilliviruses, including MeV. A common entry receptor for morbilliviruses is CD150 or signaling lymphocytic activation molecule (SLAM), which is expressed on activated lymphocytes, dendritic cell subsets, and macrophages and cells in the alveolar lumen and lining the alveolar epithelium (41–44). PoMV RNA was observed in the alveolar septa, lymphocytes of the spleen, and mononuclear leukocytes in the placenta and epicardium, which suggest that PoMV also might use CD150. Morbilliviruses infect epithelia by using nectin-4, which is expressed on the basolateral surface (45,46). We detected PoMV RNA in the allantoic epithelium of the placenta, epithelium of bronchi and bronchioles, and in rare instances the renal tubular epithelium. In addition, we detected PoMV in the endothelium of a cerebral and renal vessel, and the most extensive staining was in the cerebrum and cerebellum, including neurons. Previous studies have shown that no detectable expression of SLAM was found in human neurons (47) and extremely low expression of nectin-4 was detected in central nervous system cells and tissues (48). In contrast, CD46 is a widely distributed complement regulatory protein expressed on all nucleated cells with labeling noted in the cerebral endothelium as well as ependymal cells, neurons, and oligodendrocytes (47). Accordingly, PoMV also could use CD46 as seen in some MeV strains.
Our study provides essential information about a newly discovered pathogen within the Morbillivirus genus, but much is left to learn. The geographic distribution and species susceptibility of PoMV currently is unknown. Virus isolation to facilitate research, in vitro studies evaluating cell entry receptors, and in vivo studies to further elucidate pathogenesis and generate samples of known status for diagnostic assay development and evaluation are needed. Nonetheless, this discovery opens the door to a new and possibly more applicable model of disease to drive research forward (49).
At the time of this study, Dr. Arruda was a diagnostic pathologist at the Iowa State University Veterinary Diagnostic Laboratory and associate professor in the Department of Veterinary Diagnostic and Production Animal Medicine. Her research focused on the ecology and pathophysiology of infectious diseases of swine. She has recently taken a research position at the US Agricultural Research Services focusing on the prevention and control of influenza A virus in swine.
Acknowledgment
We thank the staff of Iowa State University Veterinary Diagnostic Laboratory for their assistance with tissue collection and processing, including Kaleigh Bell, Kevin Berkland, Sally Howard, Megan Jackson, Scott Kostohryz, Austin Kosusnik, Emily Kurtz, Haley McClure, Bonnie Snovelle, and Jerry Snyder; and the histology technicians, including Katherine Baber-Dillavou, Rosalyn Branaman, Dawn Fenneman, Victoria Green, Haley Lambert, and Linda Smith. We also thank Jennifer Groeltz-Thrush for performing the in situ hybridization assay.
References
- Rima B, Balkema-Buschmann A, Dundon WG, Duprex P, Easton A, Fouchier R, et al.; Ictv Report Consortium. ICTV virus taxonomy profile: Paramyxoviridae. J Gen Virol. 2019;100:1593–4. DOIPubMedGoogle Scholar
- Pfeffermann K, Dörr M, Zirkel F, von Messling V. Morbillivirus pathogenesis and virus-host interactions. Adv Virus Res. 2018;100:75–98. DOIPubMedGoogle Scholar
- Amarasinghe GK, Ayllón MA, Bào Y, Basler CF, Bavari S, Blasdell KR, et al. Taxonomy of the order Mononegavirales: update 2019. Arch Virol. 2019;164:1967–80. DOIPubMedGoogle Scholar
- Lamb RA, Parks GD. Paramyxoviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields virology. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2013. p. 957–95.
- Black FL. Epidemiology of Paramyxoviridae. In: Kingsbury DW, editor. The paramyxoviruses. In: Fraenkel-Conrat H and Wagner RR, series editors. The viruses. 1st ed. New York: Springer Science+Business Media; 1991. p. 509–36.
- Chiba ME, Saito M, Suzuki N, Honda Y, Yaegashi N. Measles infection in pregnancy. J Infect. 2003;47:40–4. DOIPubMedGoogle Scholar
- Furuse Y, Suzuki A, Oshitani H. Origin of measles virus: divergence from rinderpest virus between the 11th and 12th centuries. Virol J. 2010;7:52. DOIPubMedGoogle Scholar
- de Vries RD, Ludlow M, de Jong A, Rennick LJ, Verburgh RJ, van Amerongen G, et al. Delineating morbillivirus entry, dissemination and airborne transmission by studying in vivo competition of multicolor canine distemper viruses in ferrets. PLoS Pathog. 2017;13:
e1006371 . DOIPubMedGoogle Scholar - Rudd PA, Cattaneo R, von Messling V. Canine distemper virus uses both the anterograde and the hematogenous pathway for neuroinvasion. J Virol. 2006;80:9361–70. DOIPubMedGoogle Scholar
- Summers BA, Greisen HA, Appel MJ. Early events in canine distemper demyelinating encephalomyelitis. Acta Neuropathol. 1979;46:1–10. DOIPubMedGoogle Scholar
- Stephan HA, Gay GM, Ramírez TC. Encephalomyelitis, reproductive failure and corneal opacity (blue eye) in pigs, associated with a paramyxovirus infection. Vet Rec. 1988;122:6–10. DOIPubMedGoogle Scholar
- Bowden TR, Westenberg M, Wang LF, Eaton BT, Boyle DB. Molecular characterization of Menangle virus, a novel paramyxovirus which infects pigs, fruit bats, and humans. Virology. 2001;283:358–73. DOIPubMedGoogle Scholar
- Mohd Nor MN, Gan CH, Ong BL. Nipah virus infection of pigs in peninsular Malaysia. Rev Sci Tech. 2000;19:160–5. DOIPubMedGoogle Scholar
- Qiao D, Janke BH, Elankumaran S. Complete genome sequence and pathogenicity of two swine parainfluenzavirus 3 isolates from pigs in the United States. J Virol. 2010;84:686–94. DOIPubMedGoogle Scholar
- Lau SKP, Woo PCY, Wu Y, Wong AYP, Wong BHL, Lau CCY, et al. Identification and characterization of a novel paramyxovirus, porcine parainfluenza virus 1, from deceased pigs. J Gen Virol. 2013;94:2184–90. DOIPubMedGoogle Scholar
- Palinski RM, Chen Z, Henningson JN, Lang Y, Rowland RRR, Fang Y, et al. Widespread detection and characterization of porcine parainfluenza virus 1 in pigs in the USA. J Gen Virol. 2016;97:281–6. DOIPubMedGoogle Scholar
- Janke BH, Paul PS, Landgraf JG, Halbur PG, Huinker CD. Paramyxovirus infection in pigs with interstitial pneumonia and encephalitis in the United States. J Vet Diagn Invest. 2001;13:428–33. DOIPubMedGoogle Scholar
- Greig AS, Johnson CM, Bouillant AM. Encephalomyelitis of swine caused by a haemagglutinating virus. VI. Morphology of the virus. Res Vet Sci. 1971;12:305–7. DOIPubMedGoogle Scholar
- Sasahara J, Hayashi S, Kumagai T, Yamamoto Y, Hirasawa N, Munekata K, et al. On a swine virus disease newly discovered in Japan its characteristic traits of pneumonia: 1. Isolation of the virus. 2. Some properties of the virus. Virus. 1954;4:131–9. DOIGoogle Scholar
- Lipkind M, Shoham D, Shihmanter E. Isolation of a paramyxovirus from pigs in Israel and its antigenic relationships with avian paramyxoviruses. J Gen Virol. 1986;67:427–39. DOIPubMedGoogle Scholar
- Arruda B, Piñeyro P, Derscheid R, Hause B, Byers E, Dion K, et al. PCV3-associated disease in the United States swine herd. Emerg Microbes Infect. 2019;8:684–98. DOIPubMedGoogle Scholar
- Rivera-Benitez JF, García-Contreras AC, Reyes-Leyva J, Hernández J, Sánchez-Betancourt JI, Ramírez-Mendoza H. Efficacy of quantitative RT-PCR for detection of the nucleoprotein gene from different porcine rubulavirus strains. Arch Virol. 2013;158:1849–56. DOIPubMedGoogle Scholar
- Zhang J, Zheng Y, Xia XQ, Chen Q, Bade SA, Yoon KJ, et al. High-throughput whole genome sequencing of Porcine reproductive and respiratory syndrome virus from cell culture materials and clinical specimens using next-generation sequencing technology. J Vet Diagn Invest. 2017;29:41–50. DOIPubMedGoogle Scholar
- Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. DOIPubMedGoogle Scholar
- Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016;7:11257. DOIPubMedGoogle Scholar
- Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12:385. DOIPubMedGoogle Scholar
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–23. DOIPubMedGoogle Scholar
- Hunt M, Gall A, Ong SH, Brener J, Ferns B, Goulder P, et al. IVA: accurate de novo assembly of RNA virus genomes. Bioinformatics. 2015;31:2374–6. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP. Variant review with the integrative genomics viewer. Cancer Res. 2017;77:e31–4. DOIPubMedGoogle Scholar
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–9. DOIPubMedGoogle Scholar
- Gaskin JM. Canine distemper virus in domesticated cats and pigs. Adv Enzymol Relat Areas Mol Biol. 1974;40:803–6.PubMedGoogle Scholar
- Schulz C, Fast C, Schlottau K, Hoffmann B, Beer M. Neglected hosts of small ruminant morbillivirus. Emerg Infect Dis. 2018;24:2334–7. DOIPubMedGoogle Scholar
- Hernández-Jáuregui P, Ramírez Mendoza H, Mercado García C, Moreno-López J, Kennedy S. Experimental porcine rubulavirus (La Piedad-Michoacan virus) infection in pregnant gilts. J Comp Pathol. 2004;130:1–6. DOIPubMedGoogle Scholar
- Love RJ, Philbey AW, Kirkland PD, Ross AD, Davis RJ, Morrissey C, et al. Reproductive disease and congenital malformations caused by Menangle virus in pigs. Aust Vet J. 2001;79:192–8. DOIPubMedGoogle Scholar
- Philbey AW, Kirkland PD, Ross AD, Davis RJ, Gleeson AB, Love RJ, et al. An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg Infect Dis. 1998;4:269–71. DOIPubMedGoogle Scholar
- Eberhart-Phillips JE, Frederick PD, Baron RC, Mascola L. Measles in pregnancy: a descriptive study of 58 cases. Obstet Gynecol. 1993;82:797–801.PubMedGoogle Scholar
- Ali ME, Albar HM. Measles in pregnancy: maternal morbidity and perinatal outcome. Int J Gynaecol Obstet. 1997;59:109–13. DOIPubMedGoogle Scholar
- Ohyama M, Fukui T, Tanaka Y, Kato K, Hoshino R, Sugawara T, et al. Measles virus infection in the placenta of monozygotic twins. Mod Pathol. 2001;14:1300–3. DOIPubMedGoogle Scholar
- Malgarin CM, Nosach R, Novakovic P, Suleman M, Ladinig A, Detmer SE, et al. Classification of fetal resilience to porcine reproductive and respiratory syndrome (PRRS) based on temporal viral load in late gestation maternal tissues and fetuses. Virus Res. 2019;260:151–62. DOIPubMedGoogle Scholar
- Cocks BG, Chang CC, Carballido JM, Yssel H, de Vries JE, Aversa G. A novel receptor involved in T-cell activation. Nature. 1995;376:260–3. DOIPubMedGoogle Scholar
- Tatsuo H, Ono N, Yanagi Y. Morbilliviruses use signaling lymphocyte activation molecules (CD150) as cellular receptors. J Virol. 2001;75:5842–50. DOIPubMedGoogle Scholar
- Lemon K, de Vries RD, Mesman AW, McQuaid S, van Amerongen G, Yüksel S, et al. Early target cells of measles virus after aerosol infection of non-human primates. PLoS Pathog. 2011;7:
e1001263 . DOIPubMedGoogle Scholar - Mesman AW, de Vries RD, McQuaid S, Duprex WP, de Swart RL, Geijtenbeek TBH. A prominent role for DC-SIGN+ dendritic cells in initiation and dissemination of measles virus infection in non-human primates. PLoS One. 2012;7:
e49573 . DOIPubMedGoogle Scholar - Mühlebach MD, Mateo M, Sinn PL, Prüfer S, Uhlig KM, Leonard VHJ, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;480:530–3. DOIPubMedGoogle Scholar
- Noyce RS, Bondre DG, Ha MN, Lin LT, Sisson G, Tsao MS, et al. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 2011;7:
e1002240 . DOIPubMedGoogle Scholar - McQuaid S, Cosby SL. An immunohistochemical study of the distribution of the measles virus receptors, CD46 and SLAM, in normal human tissues and subacute sclerosing panencephalitis. Lab Invest. 2002;82:403–9. DOIPubMedGoogle Scholar
- Berglund L, Björling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA-K, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008;7:2019–27. DOIPubMedGoogle Scholar
- Swindle MM, Makin A, Herron AJ, Clubb FJ Jr, Frazier KS. Swine as models in biomedical research and toxicology testing. Vet Pathol. 2012;49:344–56. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: May 25, 2021
Table of Contents – Volume 27, Number 7—July 2021
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Ganwu Li and Bailey Arruda, Iowa State University Veterinary Diagnostic Laboratory, 1850 Christensen Dr, Ames, IA 50011, USA
Top