Volume 27, Number 7—July 2021
Research
Whole-Genome Analysis of Streptococcus pneumoniae Serotype 4 Causing Outbreak of Invasive Pneumococcal Disease, Alberta, Canada
Figure 2
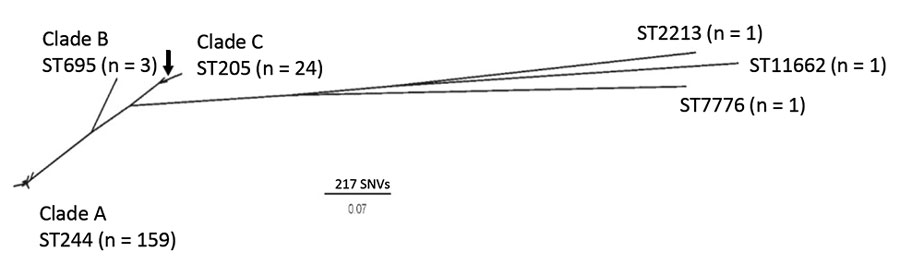
Figure 2. Maximum-likelihood core SNV unrooted phylogenetic tree of 190 Streptococcus pneumoniae serotype 4 isolates collected from patients in Alberta, Canada, 2010–2018. A total of 3,097 sites were used in the phylogeny, and 80.7% of the core genome was included. S. pneumoniae TIGR4 (arrow; National Center for Biotechnology Information accession number NC_003028.3) was used as a mapping reference. Cluster analysis did not group 1 ST205 isolate and TIGR4 with the other clade C strains. SNV, single nucleotide variant; ST, sequence type.
Page created: April 23, 2021
Page updated: June 16, 2021
Page reviewed: June 16, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.