Volume 27, Number 9—September 2021
Research
Severe Acute Respiratory Syndrome Coronavirus 2 in Farmed Mink (Neovison vison), Poland
Figure 1
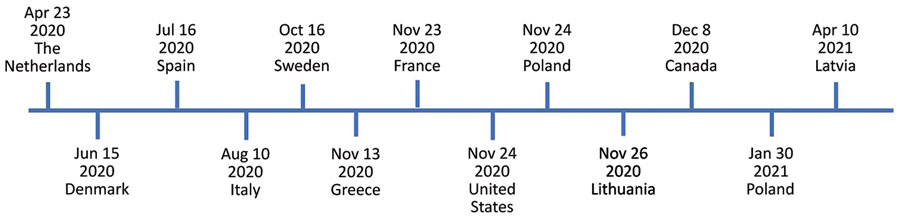
Figure 1. Timeline of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections in mink farms, Europe, according to the World Organisation for Animal Health (10). We investigated SARS-CoV-2 in mink sampled on November 24, 2020, in Pomorskie Voivodeship, northern Poland. The Polish National Veterinary Research Institute, as a national unit responsible for reporting to the World Organisation for Animal Health, detected SARS-CoV-2 infection in the same mink farm on January 30, 2021.
References
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. DOIPubMedGoogle Scholar
- Jo WK, de Oliveira-Filho EF, Rasche A, Greenwood AD, Osterrieder K, Drexler JF. Potential zoonotic sources of SARS-CoV-2 infections. Transbound Emerg Dis. 2020;10:1111.PubMedGoogle Scholar
- Mallapaty S. The search for animals harbouring coronavirus - and why it matters. Nature. 2021;591:26–8. DOIPubMedGoogle Scholar
- Alexander MR, Schoeder CT, Brown JA, Smart CD, Moth C, Wikswo JP, et al. Predicting susceptibility to SARS-CoV-2 infection based on structural differences in ACE2 across species. FASEB J. 2020;34:15946–60. DOIPubMedGoogle Scholar
- Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci U S A. 2020;117:22311–22. DOIPubMedGoogle Scholar
- Hammer AS, Quaade ML, Rasmussen TB, Fonager J, Rasmussen M, Mundbjerg K, et al. SARS-CoV-2 transmission between mink (Neovison vison) and humans, Denmark. Emerg Infect Dis. 2021;27:547–51. DOIPubMedGoogle Scholar
- Koopmans M. SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet Infect Dis. 2021;21:18–9. DOIPubMedGoogle Scholar
- Oreshkova N, Molenaar RJ, Vreman S, Harders F, Oude Munnink BB, Hakze-van der Honing RW, et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 2020;25:
200105 . DOIPubMedGoogle Scholar - Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–7. DOIPubMedGoogle Scholar
- World Organisation for Animal Health. COVID-19, 2021 [cited 2021 Jan 12]. https://www.oie.int/en/scientific-expertise/specific-information-and-recommendations/questions-and-answers-on-2019novel-coronavirus/events-in-animals
- World Health Organization. WHO-convened global study of the origins of SARS-CoV-2, 2020 [cited 2020 Dec 13]. https://www.who.int/publications/m/item/who-convened-global-study-of-the-origins-of-sars-cov-2
- Boklund A, Hammer AS, Quaade ML, Rasmussen TB, Lohse L, Strandbygaard B, et al. SARS-CoV-2 in Danish mink farms: course of the epidemic and a descriptive analysis of the outbreaks in 2020. Animals (Basel). 2021;11:164. DOIPubMedGoogle Scholar
- European Union Fur Association. Sustainable fur, 2020 [cited 2020 Dec 13]. https://www.sustainablefur.com
- Związku Przedsiębiorców i Pracodawców. Facts about breeding fur animals in Poland [in Polish]. Warsaw; 2020 [cited 2021 Jun 17]. https://zpp.net.pl/wp-content/uploads/2020/09/Raport-ZPP_Fakty-o-hodowli-zwierząt-futerkowych-w-Polsce.pdf
- European Centre for Disease Prevention and Control. COVID-19 situation update for the EU/EEA and the UK, 2021 [cited 2021 May 12]. https://www.ecdc.europa.eu/en/cases-2019-ncov-eueea
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:
2000045 . DOIPubMedGoogle Scholar - Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. DOIPubMedGoogle Scholar
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. DOIPubMedGoogle Scholar
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM, 2013 [cited 2021 May 13]. https://github.com/lh3/bwa
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. DOIPubMedGoogle Scholar
- Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–3. DOIPubMedGoogle Scholar
- Sagulenko P, Puller V, Neher RA. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:
vex042 . DOIPubMedGoogle Scholar - Sokal RR, Rohlf FJ. Biometry: the principles and practices of statistics in biological research. New York: W. H. Freeman; 1995.
- Grzybek M, Bajer A, Bednarska M, Al-Sarraf M, Behnke-Borowczyk J, Harris PD, et al. Long-term spatiotemporal stability and dynamic changes in helminth infracommunities of bank voles (Myodes glareolus) in NE Poland. Parasitology. 2015;142:1722–43. DOIPubMedGoogle Scholar
- Hemida MG, Ba Abduallah MM. The SARS-CoV-2 outbreak from a one health perspective. One Health. 2020;10:
100127 . DOIPubMedGoogle Scholar - Abdel-Moneim AS, Abdelwhab EM. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens. 2020;9:529. DOIPubMedGoogle Scholar
- Freuling CM, Breithaupt A, Müller T, Sehl J, Balkema-Buschmann A, Rissmann M, et al. Susceptibility of raccoon dogs for experimental SARS-CoV-2 infection. Emerg Infect Dis. 2020;26:2982–5. DOIPubMedGoogle Scholar
- Arya R, Kumari S, Pandey B, Mistry H, Bihani SC, Das A, et al. Structural insights into SARS-CoV-2 proteins. J Mol Biol. 2021;433:
166725 . DOIPubMedGoogle Scholar - Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. DOIPubMedGoogle Scholar
Page created: June 16, 2021
Page updated: December 08, 2021
Page reviewed: December 08, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.