Volume 27, Number 9—September 2021
Dispatch
Highly Pathogenic Avian Influenza A(H5N6) Virus Clade 2.3.4.4h in Wild Birds and Live Poultry Markets, Bangladesh
Figure 1
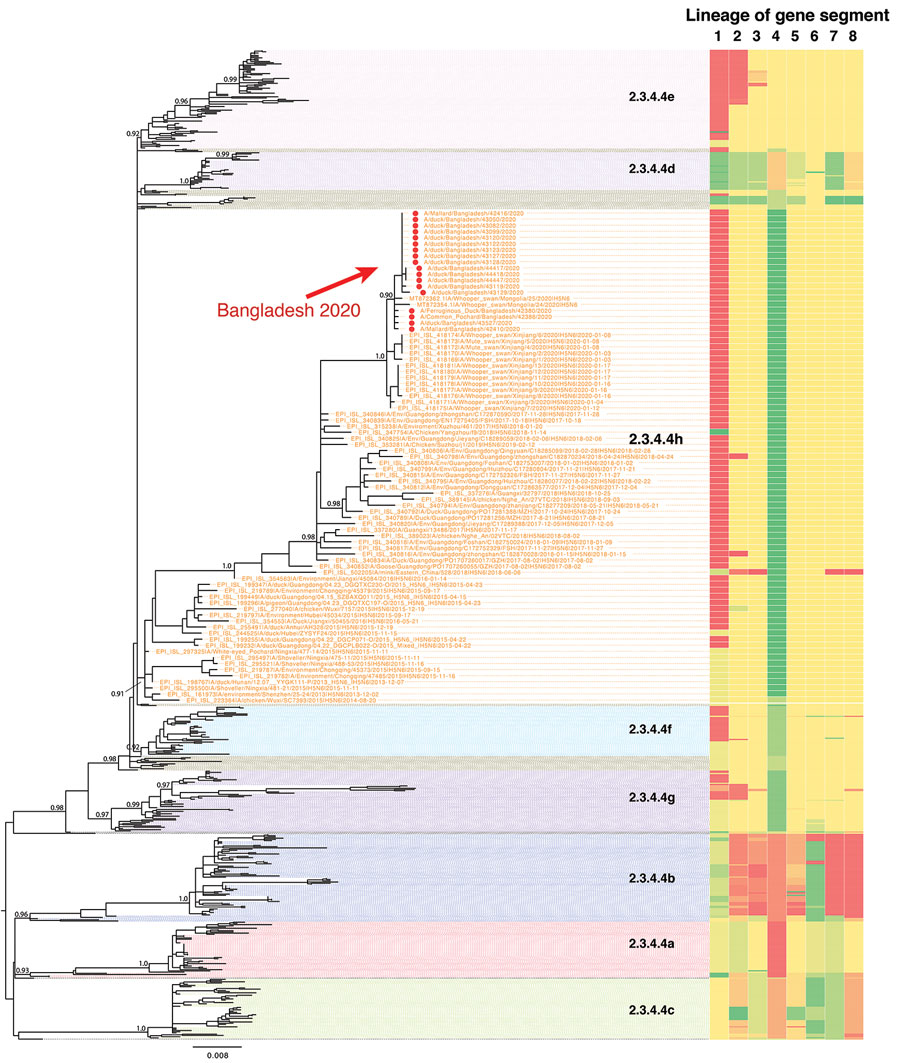
Figure 1. Phylogenetic tree of H5N6 viruses sequenced in this study, in addition to all publicly available H5N6 and closely related H5 sequences available from GenBank and GISAID. Red dots represent the Bangladesh H5N6 viruses sequenced in this study. Topological support values (SH-like support) of selected nodes are displayed.
Page created: August 10, 2021
Page updated: August 18, 2021
Page reviewed: August 18, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.