Volume 28, Number 10—October 2022
Research Letter
Emerging Tickborne Bacteria in Cattle from Colombia
Figure
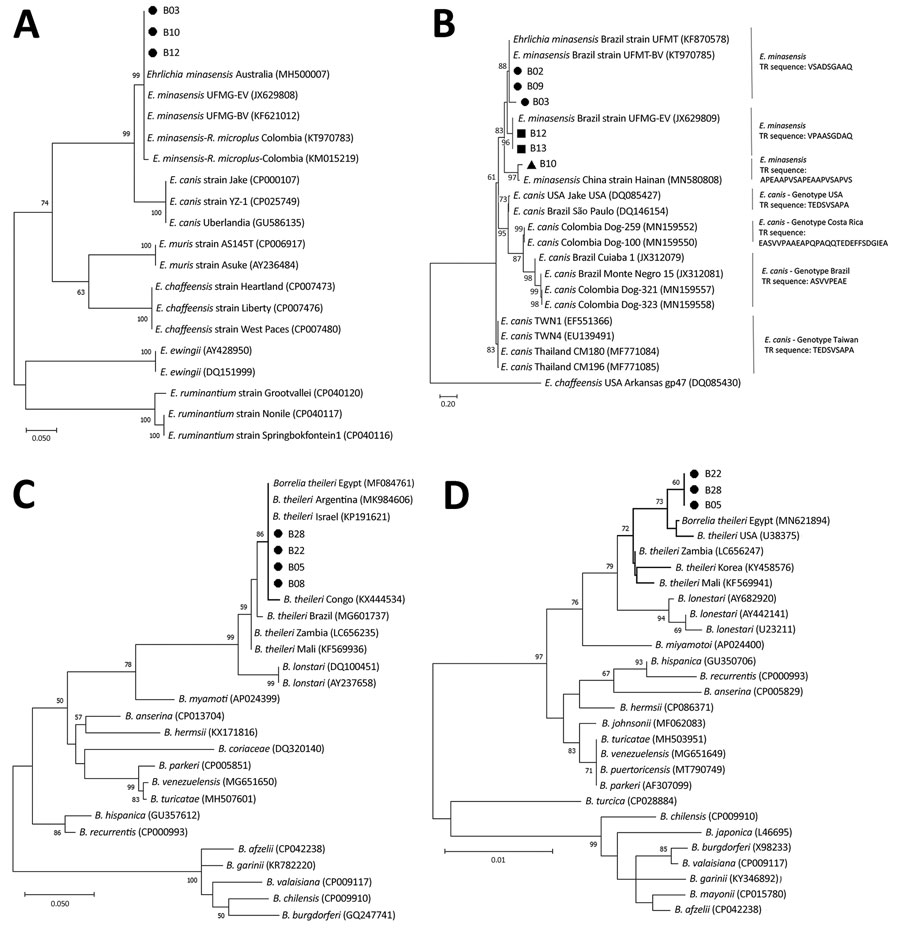
Figure. Phylogenetic analysis of emerging tickborne bacteria in cattle from Colombia. Phylogenetic trees are shown for dsb genes (A), Trp36 proteins (amino acid sequences) (B), flaB genes (C), and 16S rRNA genes (D). We performed PCR on blood samples from cattle in El Tambo and Santander de Quilichao, Cauca department, Colombia to detect dsb and trp36 genes for Ehrlichia sp.; flaB and 16S rRNA genes for Borrelia sp.; and rpoB, msp4, and msp1a genes for Anaplasma sp. We generated phylogenetic trees using the maximum-likelihood method and Tamura 3-parameter model with a gamma distribution parameter of 0.28 to compare evolutionary relationships between our sequences and publicly available sequences from Genbank (accession numbers indicated on trees). We applied bootstrap tests using 1,000 replicates; bootstrap values are shown at key nodes. The ●, ■, and ▲ symbols represent the differential clustering of sequences obtained in this study. Scale bars indicate nucleotide substitutions per site.