Volume 28, Number 11—November 2022
Dispatch
Polyclonal Dissemination of OXA-232 Carbapenemase–Producing Klebsiella pneumoniae, France, 2013–2021
Figure 2
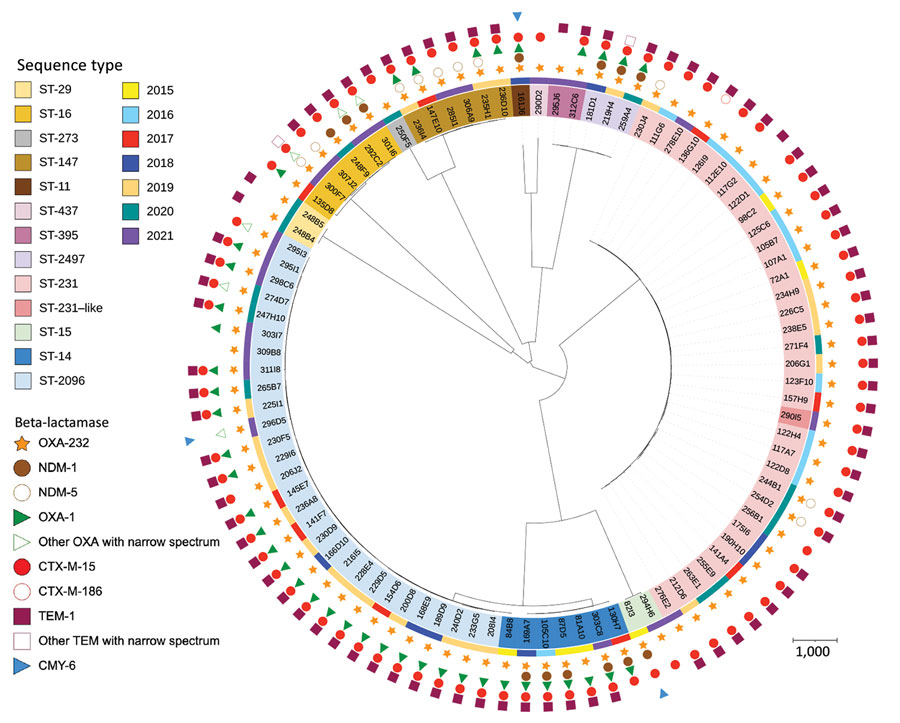
Figure 2. Phylogenetic relationship of OXA-232–producing K. pneumoniae ST-231 (A) and ST-2096 (B) analyzed at the National Reference Center for Carbapenem-Resistant Enterobacterales, France 2013–2021.The phylogenetic trees were built with an SNP analysis approach. Scale bars under trees indicate the number of SNPs per position of common sequences. OXA, oxacillinase; SNP, single nucleotide polymorphism; ST, sequence type.
Page created: August 17, 2022
Page updated: October 24, 2022
Page reviewed: October 24, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.