Volume 28, Number 2—February 2022
Research Letter
Spillover of Canine Parvovirus Type 2 to Pigs, South Dakota, USA, 2020
Cite This Article
Citation for Media
Abstract
In 1978, canine parvovirus type 2 originated from spillover of a feline panleukopenia–like virus, causing a worldwide pandemic of enteritis and myocarditis among canids. In 2020, the virus was identified in pigs in South Dakota, USA, by PCR, sequencing, in situ hybridization, and serology. Genetic analysis suggests spillover from wildlife.
Canine parvovirus type 2 (CPV-2) is a variant of the species Carnivore protoparvovirus 1, which can cause severe disease in carnivores of many species (1–3). Besides CPV-2, which causes enteritis in dogs of all ages and myocarditis in puppies, the virus species includes feline panleukopenia virus, which causes severe enteritis and leukopenia in cats of all ages (4). In 1978, CPV-2 emerged and caused a worldwide pandemic after spillover from a feline panleukopenia virus–like virus in wildlife. Subsequent adaptation to canine hosts led to genetic and antigenic diversification into subtypes 2a, 2b, and 2c (5). Continued CPV host switching has been documented; spillover to wildlife (including skunks, raccoons, coyotes) has resulted in clinical disease and asymptomatic infection (2).
In October 2020, a dead pig was submitted to South Dakota State University (Brookings, SD, USA) for diagnostic testing. Histopathologic examination revealed mild to moderate enteritis, hepatitis, and visceral edema. Hemolytic Escherichia coli was isolated. No significant lung lesions were noted. Approximately 8 months later, we performed viral metagenomic sequencing on archived lung tissue for an unrelated research project and unexpectedly identified CPV-2. Using a S′ nuclease PCR (Integrated DNA Technologies, https://www.idtdna.com), we confirmed that the sample was CPV-2 positive; cycle threshold (Ct) was 24.4. Sanger sequencing of overlapping amplicons confirmed the CPV-2 genome sequence determined by metagenomic sequencing. We submitted the strain SDS21601 sequence to GenBank (accession no. MZ666397).
We used a S′ nuclease PCR to test 90 archived porcine lung samples submitted for respiratory disease diagnostic testing for CPV-2. Of the 90 samples, 9 (10%) were positive for CPV-2, including those with strain SDS21601, and Ct values were 22.4–36.3. The samples were collected September–November 2020 from swine farms within 150 miles of Brookings. We sequenced the genome from a second strongly positive sample (Ct 22.4) and submitted strain SDS21608 to GenBank (accession no. MZ666398). An amplicon from 4 of the remaining 7 samples positive by PCR was generated by PCR and confirmed as CPV-2 by Sanger sequencing. The 3 samples that failed to yield a CPV-2–specific amplicon had Ct values >32. Sequence comparison showed 99.9% nt identity between SDS21601 and SDS21608. blastp (https://blast.ncbi.nlm.nih.gov) analysis of SDS21601 virus capsid protein (VP) 2 found 100% identity to CPV-2 from a coyote sampled in Montana in 2012. Analysis of the VP2 amino acid sequences identified an F212I mutation previously identified only from US wildlife, mainly coyotes.
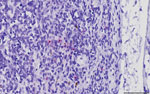
We performed in situ hybridization on archived formalin-fixed paraffin-embedded tissues from SDS21608 by using a commercially available CPV-2 probe. CPV-2 nucleic acids were hybridized sporadically as intracytoplasmic punctate signals in few monocyte–macrophage lineage cells in the medullary and subcapsular sinuses of a bronchial lymph node (Figure). However, the primary anatomic site of CPV-2 infection and replication was not determined. In other examined tissues, we observed neither typical virus-associated microscopic lesions as seen in carnivores nor obvious CPV-2 hybridization signals.
To further investigate the extent of CPV-2 circulation among swine, 8 months after collection of the CPV-2–positive lung tissue, we collected 20 serum samples from multiparous sows on the farm where strain SDS21601 originated. Of the 20 samples, 13 (65%) were positive for CPV-2–specific antibodies by hemagglutination inhibition (HI) assay; titers were 10–40 (Table). Nearly all serum samples (19 of 20) had antibody titers to porcine parvovirus 1 (PPV-1), ranging from 16 to 4,096. This result was expected given that pigs on the farm received commercial PPV-1 vaccine before farrowing. There was no correlation between CPV-2 and PPV-1 HI titers, indicating a lack of cross-reactivity between CPV-2 and PPV-1 antibodies in the HI assay.
To further investigate seroprevalence of CPV-2 in South Dakota, we randomly selected 25 sow serum samples from unrelated submissions collected at 5 farms (5 samples/farm) and analyzed them by HI for CPV-2. Of the 25 samples, 23 (92%) were positive for CPV-2; titers were 10–80. Together with the 10% positivity detected by quantitative PCR, these results suggest widespread CPV-2 infection of swine in South Dakota.
Members of Carnivore protoparvovirus 1 display >98% identity. Amino acid residue 300 of VP2 has been shown to be a critical determinant for the cross-species transfer of CPV-2 between carnivores of different species (6). Glycine 300 and tyrosine 305, observed in the VP2 of both swine CPV-2 strains (SDS21601 and SDS21608), are diagnostic of CPV-2 isolates from canids (7). The F212I mutation present in both swine CPV-2 strains, which was previously found only in wildlife, suggests a sylvatic origin. Of the species in which F212I has been identified, only coyotes are common in the agricultural areas of the upper US Midwest and are peridomestic. We hypothesize that the source of swine CPV-2 infection is CPV-2–positive coyote feces.
Our results demonstrate spillover of CPV-2 to swine. CPV-2 has been associated with severe enteritis in insectivorous Taiwanese pangolin (Manis pentadactyla pentadactyla), further demonstrating the propensity of CPV-2 to overcome host barriers (8). The ability of CPV-2 to cause disease in swine remains unknown; further surveillance is warranted because this spillover may threaten the health of swine herds.
Dr. Temeeyasen is a research associate at the Animal Disease Research and Diagnostic Laboratory, South Dakota State University, in Brookings. His primary research interest is pathogenic porcine and bovine viruses.
Acknowledgments
We thank Martha Ohnstad and Craig Long for assistance with metagenomic sequencing.
This project was in part funded by start-up funds provided to B.H. by the South Dakota State University Agricultural Experiment Station Hatch and Animal Health funds in addition to the South Dakota State University Center for Biologics Research and Commercialization.
References
- Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, et al. The family Parvoviridae. Arch Virol. 2014;159:1239–47. DOIPubMedGoogle Scholar
- Allison AB, Kohler DJ, Fox KA, Brown JD, Gerhold RW, Shearn-Bochsler VI, et al. Frequent cross-species transmission of parvoviruses among diverse carnivore hosts. J Virol. 2013;87:2342–7. DOIPubMedGoogle Scholar
- Steinel A, Parrish CR, Bloom ME, Truyen U. Parvovirus infections in wild carnivores. J Wildl Dis. 2001;37:594–607. DOIPubMedGoogle Scholar
- Parrish CR. Pathogenesis of feline panleukopenia virus and canine parvovirus. Baillieres Clin Haematol. 1995;8:57–71. DOIPubMedGoogle Scholar
- Wasik BR, de Wit E, Munster V, Lloyd-Smith JO, Martinez-Sobrido L, Parrish CR. Onward transmission of viruses: how do viruses emerge to cause epidemics after spillover? Philos Trans R Soc Lond B Biol Sci. 2019;374:
20190017 . DOIPubMedGoogle Scholar - Allison AB, Organtini LJ, Zhang S, Hafenstein SL, Holmes EC, Parrish CR. Single mutations in the VP2 300 loop region of the three-fold spike of the carnivore parvovirus capsid can determine host range. J Virol. 2015;90:753–67. DOIPubMedGoogle Scholar
- Allison AB, Kohler DJ, Ortega A, Hoover EA, Grove DM, Holmes EC, et al. Host-specific parvovirus evolution in nature is recapitulated by in vitro adaptation to different carnivore species. PLoS Pathog. 2014;10:
e1004475 . DOIPubMedGoogle Scholar - Chang YC, Lin ZY, Lin YX, Lin KH, Chan FT, Hsiao ST, et al. Canine parvovirus infections in Taiwanese pangolins (Manis pentadactyla pentadactyla). Vet Pathol. 2021;58:743–50. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: January 13, 2022
Table of Contents – Volume 28, Number 2—February 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Ben M. Hause, Department of Veterinary and Biomedical Sciences, South Dakota State University, Box 2175, 1155 North Campus Dr, Brookings, SD 57007, USA
Top