Volume 28, Number 2—February 2022
Dispatch
Neisseria gonorrhoeae FC428 Subclone, Vietnam, 2019–2020
Figure
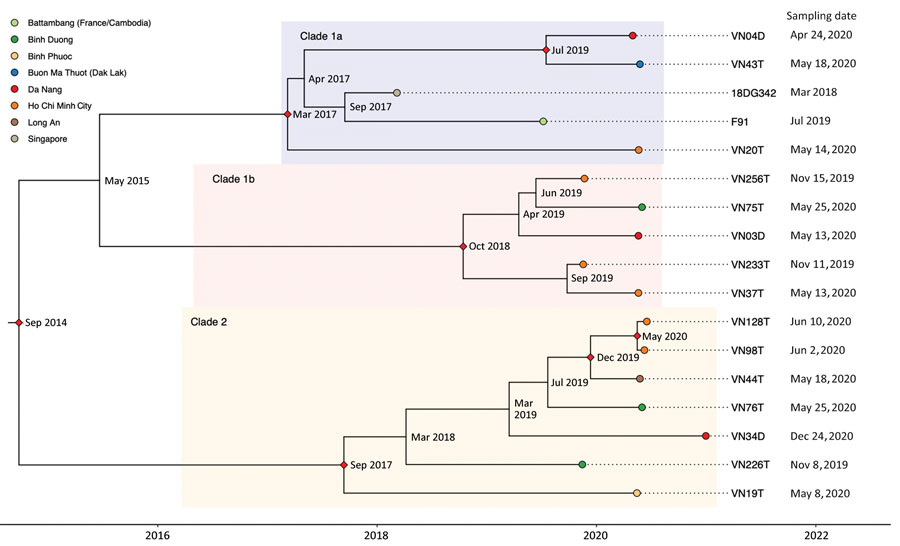
Figure. Time-scaled Bayesian maximum clade credibility phylogenetic tree of Neisseria gonorrhoeae ST13871 (17 isolates) with date of collection and location of collected isolates, Vietnam, 2019–2020. Red diamonds show posterior probability >90%; internal node labels show estimated time to most recent common ancestor.
1These first authors contributed equally to this article.
Page created: December 02, 2021
Page updated: January 22, 2022
Page reviewed: January 22, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.