Volume 28, Number 3—March 2022
Dispatch
Encephalitozoon cuniculi and Extraintestinal Microsporidiosis in Bird Owners
Figure
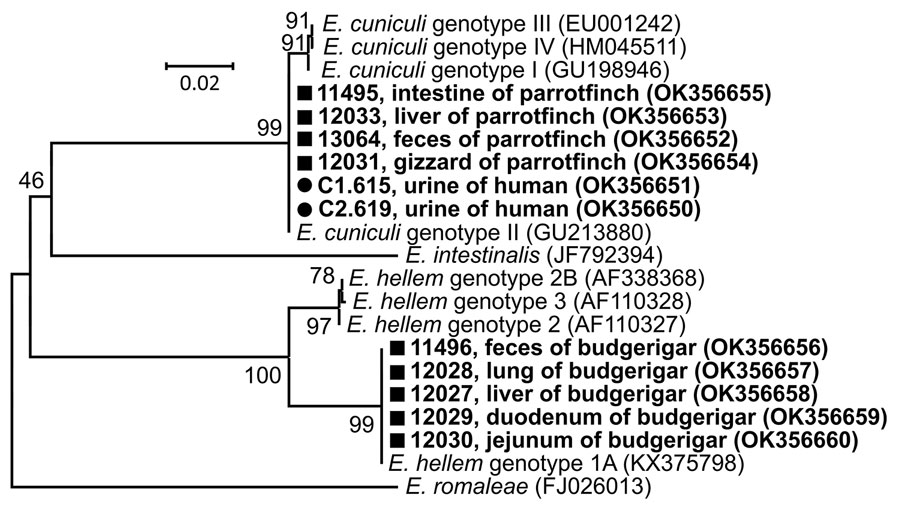
Figure. Phylogenetic relationships of Encephalitozoon cuniculi genotype II and E. hellem genotype 1A obtained from 2 exotic bird breeders and 2 of their birds compared with other Encephalitozoon species and genotypes. Bold type indicates sequences obtained in this study, identified by isolate number (e.g., C1.615); black circles indicate isolates from humans; squares indicate isolates from birds. We analyzed a partial sequence of 16S rRNA gene, the entire internal transcribed spacer region, and a partial sequence of 5.8S rRNA gene inferred by neighbor-joining analyses and computed using the Tamura 3-parameter method. We modeled the rate variation among sites with a gamma distribution. Percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The final dataset contained a total of 220 positions. GenBank accession numbers are in parentheses. Scale bar indicates nucleotide substitutions per site.