Volume 28, Number 4—April 2022
Synopsis
Phylogenetic Analysis of Spread of Hepatitis C Virus Identified during HIV Outbreak Investigation, Unnao, India
Figure 3
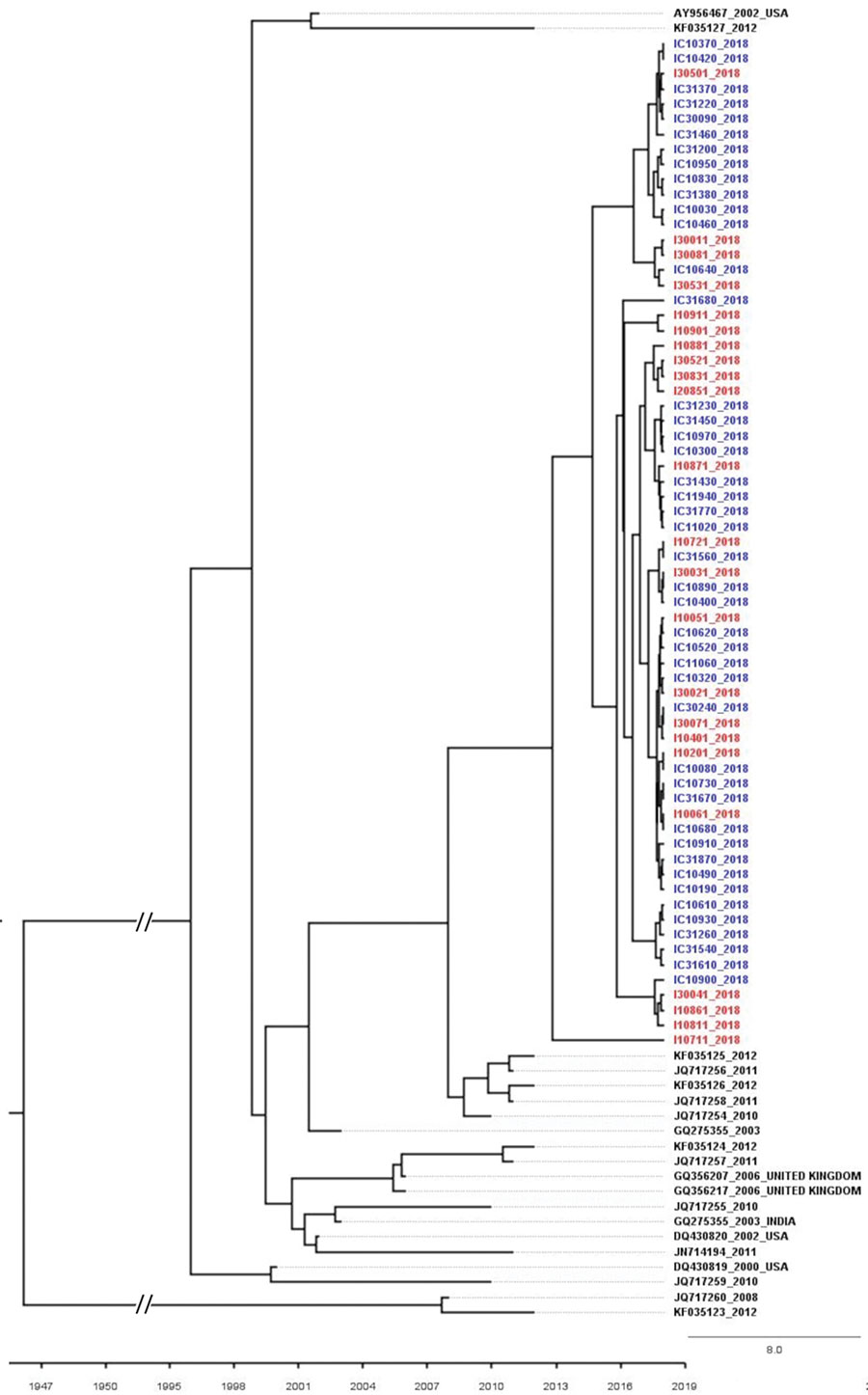
Figure 3. Time to most recent common ancestor estimated using hepatitis C virus (HCV) NS5B gene sequences of isolates from HCV antibody–positive persons identified during HIV outbreak investigation, Unnao, India. Red indicates samples with HIV–HCV coinfection; blue indicates samples with HCV monoinfection. Mean estimated time to most recent common ancestor of HCV isolates with the respective 95% highest posterior density interval is 2012 (2008–2014), with posterior probability value of 1. GenBank accession numbers are provided for reference sequences. Scale bar indicates branch lengths. I, HIV–HCV co-infection; IC, HCV monoinfection.
Page created: February 15, 2022
Page updated: March 21, 2022
Page reviewed: March 21, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.