Volume 28, Number 4—April 2022
Dispatch
In Vitro Confirmation of Artemisinin Resistance in Plasmodium falciparum from Patient Isolates, Southern Rwanda, 2019
Abstract
Artemisinin resistance in Plasmodium falciparum is conferred by mutations in the kelch 13 (K13) gene. In Rwanda, K13 mutations have increased over the past decade, including mutations associated with delayed parasite clearance. We document artemisinin resistance in P. falciparum patient isolates from Rwanda carrying K13 R561H, A675V, and C469F mutations.
Artemisinin-based combination therapies (ACTs) have contributed greatly to the global decline of illness and death from malaria (1). However, the novel emergence of artemisinin resistance in eastern Africa has threatened the effectiveness of these breakthrough treatments (2–4). To avert potential disaster resulting from increased resistant malaria cases, the nature and extent of this resistance in Africa urgently needs to be characterized.
Artemisinin resistance is conferred by some Plasmodium falciparum kelch 13 (K13) gene mutations, only a few of which are validated markers of resistance, defined by both in vitro resistance and delayed parasite clearance in treated patients. For candidate markers, only parasite clearance applies (1). In Rwanda, K13 mutations have increased over the past decade. K13 R561H, a validated marker associated with delayed parasite clearance, was recently observed in >10% of P. falciparum–positive samples (2,3,5). In neighboring Uganda, artemisinin resistance conferred by another mutation, K13 A675V, has recently been reported (4). We document in vitro artemisinin resistance in 3 P. falciparum patient isolates from Rwanda carrying K13 R561H, A675V, and C469F mutations.
We recruited malaria patients in Huye District, Rwanda, during September–December 2019 and documented patient characteristics and consent, ethical clearance, and K13 variants elsewhere (2). Within 6 hours of sample collection, we cryopreserved all 66 P. falciparum isolates in ethylenediaminetetraacetic acid by washing the red blood cell pellet, adding freezing solution (3% sorbitol, 28% glycerol, 0.65% NaCl), and freezing at −80°C. Eight of the 66 isolates carried nonsynonymous K13 mutations (2). We successfully thawed and culture-adapted 4 of the isolates in which we identified K13 mutations: R561H, the current prevalent mutation in Rwanda; A675V, found in 11% of P. falciparum samples in Uganda; C469F, another candidate marker; and V555A, which is of unknown significance.
We conducted a 0–3-h postinvasion ring-stage susceptibility assay (RSA) with the active metabolite dihydroartemisinin (6). We exposed ring stages to a 6-h pulse of 700 nmol/L dihydroartemisinin and cultured exposed and nonexposed isolates in vitro in triplicate for 72 h. We counted parasite density per ≥10,000 red blood cells on Giemsa-stained thin blood films and calculated the means of triplicates. Dividing parasite density in dihydroartemisinin-exposed cultures by the density in nonexposed cultures provided the RSA survival rate. We considered results if 72-h growth rates exceeded 1.5× rates in the nonexposed controls and had >3 successful independent triplicate experiments per isolate. We also assessed 50% inhibitory concentrations (IC50) (7). We exposed synchronized ring-stage parasites for 72 h across a range of dihydroartemisinin concentrations (0–1 µmol/L) in duplicate or triplicate and in ≥3 independent experiments. We measured growth by SYBR Green I staining (ThermoFisher, https://www.thermofisher.com) and performed photometric assessment using FilterMax F5 microplate readers (Molecular Devices, https://www.moleculardevices.com). We estimated IC50 using a 4-parameter fit dose-response curve. For artemisinin-susceptible parasites, 2 cultured wild-type isolates from patients in Rwanda grew too poorly for RSA and IC50 assays and were replaced by artemisinin-susceptible K13 wild-type strain NF54, which is of putative African origin. We assayed isolates in parallel with NF54 and compared IC50 by Student t-test. We performed analyses using R version 3.6.3, including the drc (dose-response curve) package (https://cran.r-project.org/web/packages/drc/drc.pdf).
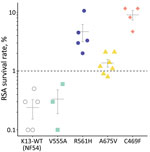
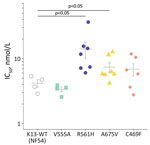
RSAs yielded mean ± SE survival rates of 0.2% ± 0.1% for the NF54 strain and 0.3% ± 0.1% for V555A, well below the World Health Organization–accepted 1% resistance threshold (1,6). In contrast, 3 other isolates with K13 mutations had >1% mean survival rates: 4.7% ± 1.5% for R561H (prevalent in Rwanda), 1.4% ± 0.2% for A675V (prevalent in Uganda), and 9.0% ± 1.6% for C469F (Figure 1). Conventional susceptibility testing yielded mean IC50 of 4.2 ± 0.5 nmol/L for dihydroartemisinin for the NF54 strain and 3.4 ± 0.3 nmol/L for V555A. IC50 levels were higher in isolates with dihydroartemisinin-resistant RSA findings: 14.1 ± 4.0 nmol/L for R561H, 7.4 ± 3.3 nmol/L for A675V, and 6.9 ± 1.5 nmol/L for C469F (Figure 2).
We determined the regional origin of the 4 tested patient isolates by single-nucleotide polymorphism (SNP) barcoding. We typed 23 SNPs to group into haplotypes associated with geographic origin (7,8). The R561H isolate displayed haplotype 9 and the other isolates haplotype 22 (9), confirming African ancestry.
Artemisinin resistance is defined by RSA results and delayed parasite clearance in treated patients. In Africa, abundant K13 variants circulate, but very few have been defined in terms of drug susceptibility (1). The K13 mutation R561H, which has emerged in Rwanda (2,5), confers delayed parasite clearance (3). We found that a patient isolate with the R561H mutation from Rwanda was in vitro artemisinin resistant. Taken together, these results strongly suggest that R561H is a marker of resistance in Rwanda, a finding that needs to be confirmed in larger sample-size research. The same need for confirmation applies to K13 candidate resistance markers A675V, recently characterized in Uganda (4), and C469F (1).
RSA survival rates for K13 R561H P. falciparum in our study concord with levels in multiple gene-edited P. falciparum lines (5,10). Also in line with our findings are high RSA survival rates in A675V isolates from neighboring Uganda, where K13 A675V was found in 11% and C469Y (but not C469F) in 2% of P. falciparum isolates collected during 2017–2019. Both mutations are associated with delayed parasite clearance (4). Isolates with increased survival rates also showed higher dihydroartemisinin IC50 levels. If this association is confirmed, IC50 assays that are much less labor-intensive could be useful for flagging isolates deserving additional testing by RSA.
The small number of isolates we evaluated was an obvious limitation of our study. Ideally, we would have compared the effects of individual mutations in wild-type isolates from Rwanda with study isolates, but the few selected performed poorly in vitro and were replaced by the artemisinin-sensitive NF54 strain, enabling us to verify that the RSA was working properly. A study strength is the detailed characterization of the susceptibility and ancestry of isolates.
RSA data on suspicious K13 isolates from Africa are scarce but essential and urgent for the situational evaluation of artemisinin resistance emerging in Africa. K13 mutations have conferred a wide range of artemisinin susceptibility when introduced in different parasite lines (10). Of note, artemisinin resistance identified in Rwanda and Uganda is of indigenous origin, not imported from Asia where resistance has been prevalent for years (1). These 2 observations argue for the need for local characterization of artemisinin resistance in circulating parasites.
Artemisinin resistance alone does not necessarily lead to ACT treatment failure, and efficacy in Rwanda still is high (3). However, resistance leaves the partner drug unprotected, potentially leading to resistance developing to that component as well. Eventually, this process could result in increased ACT treatment failure, which has already been observed in southeast Asia (11,12). In Africa, this development might be delayed because of prevalent partial immunity contributing to parasite elimination and high transmission increasing the likelihood of resistance allele outcrossing. Nonetheless, in Rwanda, where artemether/lumefantrine is the first-line antimalarial drug combination, a shift in the P. falciparum multidrug resistance 1 (pfmdr1) genotype pattern over the past decade suggests an increasingly lumefantrine-tolerant phenotype (13,14), although pfmdr1 is not a validated marker for lumefantrine resistance.
Recent research indicates that the R561H mutation is fitness neutral (10), implying its wider dissemination even without drug pressure. So far, a viable alternative to ACTs is not in sight. Increasing resistance, combined with the lack of effective alternative antimicrobial drugs, suggests a pessimistic scenario for sub-Saharan Africa, considering the region’s high malaria burden. Large-scale monitoring, containment strategies, and early consideration of 3-drug ACTs (15) are required to control widespread artemisinin resistance in Africa.
Ms. van Loon is a PhD candidate at the Institute of Tropical Medicine and International Health, Charité Universitaetsmedizin Berlin. She is interested in infectious disease epidemiology and public health. This manuscript forms part of her PhD thesis.
Acknowledgments
We are grateful to the staff of Sovu Health Centre and Kabutare District Hospital for their collaboration and help. We thank Megan Peedell and Katja Puestow for their support in the laboratory.
The German Research Foundation financially supported this study through grants to W.v.L. (GRK2046) and R.O. and C.B. (GRK2290). The funding bodies had no role in designing the study, collecting, analyzing, or interpreting data, or writing the manuscript.
References
- World Health Organisation Global Malaria Programme. Report on antimalarial drug efficacy, resistance and response: 10 years of surveillance (2010–2019). 2020 [cited 2021 Nov 1]. https://apps.who.int/iris/rest/bitstreams/1316082/retrieve
- Bergmann C, van Loon W, Habarugira F, Tacoli C, Jäger JC, Savelsberg D, et al. Increase in kelch 13 polymorphisms in Plasmodium falciparum, southern Rwanda. Emerg Infect Dis. 2021;27:294–6. DOIPubMedGoogle Scholar
- Uwimana A, Umulisa N, Venkatesan M, Svigel SS, Zhou Z, Munyaneza T, et al. Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: an open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect Dis. 2021;21:1120–8. DOIPubMedGoogle Scholar
- Balikagala B, Fukuda N, Ikeda M, Katuro OT, Tachibana SI, Yamauchi M, et al. Evidence of artemisinin-resistant malaria in Africa. N Engl J Med. 2021;385:1163–71. DOIPubMedGoogle Scholar
- Uwimana A, Legrand E, Stokes BH, Ndikumana JM, Warsame M, Umulisa N, et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. [Erratum in: Nat Med. 2021;27:1113–5.]. Nat Med. 2020;26:1602–8. DOIPubMedGoogle Scholar
- Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis. 2013;13:1043–9. DOIPubMedGoogle Scholar
- Machado M, Murtinheira F, Lobo E, Nogueira F. Whole-cell SYBR Green I assay for antimalarial activity assessment. Ann Clin Med Microbio. 2016;2:1010.
- Preston MD, Campino S, Assefa SA, Echeverry DF, Ocholla H, Amambua-Ngwa A, et al. A barcode of organellar genome polymorphisms identifies the geographic origin of Plasmodium falciparum strains. Nat Commun. 2014;5:4052. DOIPubMedGoogle Scholar
- Hu F, Zou Q, Li Y, Zhu G, Zhou H, Zhang M, et al. A PCR-based technique to track the geographic origin of Plasmodium falciparum with 23-SNP barcode analysis. Front Public Health. 2021;9:
649170 . DOIPubMedGoogle Scholar - Stokes BH, Dhingra SK, Rubiano K, Mok S, Straimer J, Gnädig NF, et al. Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness. eLife. 2021;10:
e66277 . DOIPubMedGoogle Scholar - Phyo AP, Ashley EA, Anderson TJC, Bozdech Z, Carrara VI, Sriprawat K, et al. Declining efficacy of artemisinin combination therapy against P. falciparum malaria on the Thai–Myanmar border (2003–2013): the role of parasite genetic factors. Clin Infect Dis. 2016;63:784–91. DOIPubMedGoogle Scholar
- van der Pluijm RW, Imwong M, Chau NH, Hoa NT, Thuy-Nhien NT, Thanh NV, et al. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis. 2019;19:952–61. DOIPubMedGoogle Scholar
- van Loon W, Bergmann C, Habarugira F, Tacoli C, Savelsberg D, Oliveira R, et al. Changing pattern of Plasmodium falciparum pfmdr1 gene polymorphisms in southern Rwanda. Antimicrob Agents Chemother. 2021;65:
e0090121 . DOIPubMedGoogle Scholar - Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnädig N, Uhlemann AC, et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun. 2016;7:11553. DOIPubMedGoogle Scholar
- Tindana P, de Haan F, Amaratunga C, Dhorda M, van der Pluijm RW, Dondorp AM, et al. Deploying triple artemisinin-based combination therapy (TACT) for malaria treatment in Africa: ethical and practical considerations. Malar J. 2021;20:119. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: March 08, 2022
Table of Contents – Volume 28, Number 4—April 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Welmoed van Loon, Institute of Tropical Medicine and International Health, Charité-Universitaetsmedizin Berlin, Campus Virchow-Klinikum, Augustenburger Platz 1, 13353 Berlin, Germany
Top