Volume 28, Number 4—April 2022
Research Letter
Amplification Artifact in SARS-CoV-2 Omicron Sequences Carrying P681R Mutation, New York, USA
Figure
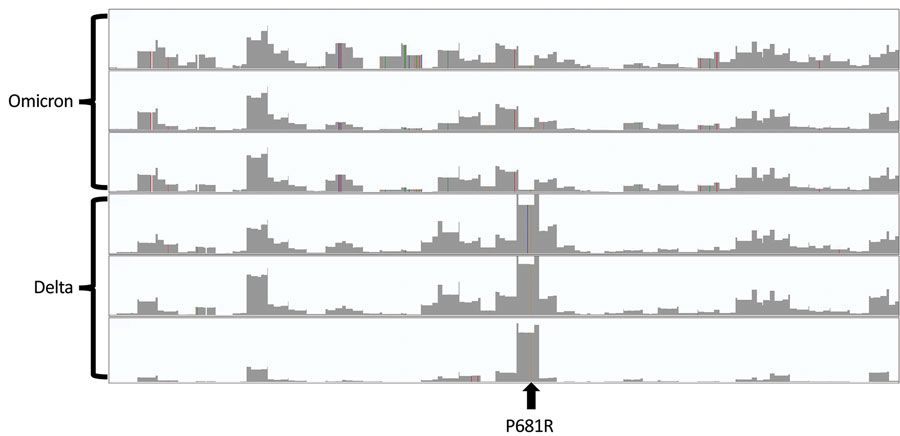
Figure. Integrated Genome Viewer (https://software.broadinstitute.org) plots of sequencing coverage of the entire length of the spike gene (3,821 nt, x-axis) of 3 representative sequences of the Omicron variant and 3 representative sequences of the Delta variant of severe acute respiratory syndrome coronavirus 2, processed in the same batch. The y-axis for each sequence is 250,000 reads. Although coverage plots are basically identical for other regions of the spike gene, coverage of the P681 position is the highest for Delta and one of the lowest for Omicron.
Page created: January 26, 2022
Page updated: March 19, 2022
Page reviewed: March 19, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.