Volume 28, Number 5—May 2022
Dispatch
Rapid Replacement of SARS-CoV-2 Variants by Delta and Subsequent Arrival of Omicron, Uganda, 2021
Abstract
Genomic surveillance in Uganda showed rapid replacement of severe acute respiratory syndrome coronavirus 2 over time by variants, dominated by Delta. However, detection of the more transmissible Omicron variant among travelers and increasing community transmission highlight the need for near–real-time genomic surveillance and adherence to infection control measures to prevent future pandemic waves.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiologic agent of human coronavirus disease (COVID-19), which was declared by the World Health Organization to be a global pandemic in March 2020 (1). Since the beginning of the pandemic, COVID-19 has caused enormous socioeconomic destruction (2) and has resulted in >5 million deaths worldwide.
A study conducted by the Medical Research Council/Uganda Virus Research Institute (MRC/UVRI) and the London School of Hygiene and Tropical Medicine (LSHTM) Uganda Research Unit (Entebbe, Uganda) during the early phase of the pandemic showed that most SARS-CoV-2 infections were imported and consisted of several lineages that included A, B, B.1, B.1.1, B.1.1.1, and B.4 (3). A subsequent study that covered the period from December 2020 through January 2021 showed that a SARS-CoV-2 lineage A variant (A.23.1) had emerged and become the dominant variant in Uganda (4).
The UVRI and its partners, such as the MRC/UVRI and LSHTM, contribute to the SARS-CoV-2 response in Uganda. As part of routine national genomic surveillance, we identified circulating variants during June‒December 2021 and analyzed trends of SARS-CoV-2 lineages over time.
We conducted SARS-CoV-2 whole-genome deep sequencing for 266 nasooropharyngeal samples collected during June‒December 2021 from 28 travelers arriving at Entebbe International Airport and from 238 patients in Uganda from 18 districts (Kampala, Wakiso, Mpigi, Kalungu, Kalangala, Dokolo, Amudat, Moroto, Kassanda, Gulu, Arua, Koboko, Amuru, Lamwo, Kwania, Apac, Kisoro, and Mityana). All samples had tested positive for SARS-CoV-2 by reverse transcription PCR with cycle threshold values <30 and were sequenced by using Illumina MiSeq (https://www.illumina.com) (n = 236, 88.7%) and Oxford Nanopore MinION (https://nanoporetech.com) (n = 30, 11.3%) next-generation sequencing platforms. Most (77%) samples sequenced were from the central region of Uganda (mostly from Kampala, Wakiso, Mpigi, and Kalungu); fewer samples came from the northern (13.9%) and western regions (8.2%) of the country.
We assembled deep sequence reads by using the genome detective software (5) (for the Illumina MiSeq‒generated sequence reads) and Nanopolish/Medaka (https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html) (for the Nanopore‒generated sequence reads) to obtain high-quality SARS-CoV-2 genomes with >80% coverage. We performed quality control of all sequences to check for adequate coverage, indels, and frameshifts. We performed mutation calling by using Nextclade (https://clades.nextstrain.org), followed by SARS-CoV-2 lineage analysis with Pangolin (https://github.com/cov-lineages/pangolin). To analyze trends of SARS-CoV-2 lineages over time, we downloaded all sequences from Uganda in GISAID (https://www.gisaid.org) (950 sequences as of January 10, 2022).
Results showed that most (195, 73.3%) of the 266 SARS-CoV-2 sequences genotyped were the Delta variant (B.1.617.2 and other AY.1, AY.4, AY.33, AY.39, AY.46, AY.46.4 sublineages), a variant of concern (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants; accessed January 10, 2022). Another variant of concern we identified was the Omicron variant (B.1.1.529 and BA.1 sublineage) (28, 10.5%). We also identified the Eta variant (B.1.525) (2, 0.8%) and other variants (41, 15.4%) mostly of the A and B lineages (Figure 1).
Uganda is in the third wave of the COVID-19 pandemic (Figure 2, panel A). During the first wave (December 2020‒January 2021), the A.23.1 variant dominated (4). During second wave (May‒July 2021) and by June 2021, Delta dominated all variants reported. We report the numbers and percentage of SARS-CoV-2 genomes generated and variants reported over time based on 950 sequences from Uganda deposited in GISAID (Figure 2, panels B, C). The first Kappa variant (B.1.617.1) was identified in March 2021. However, in June 2021, the Delta variant reached its peak and comprised >90% of all circulating variants. SARS-CoV-2 variants previously reported (3,4) have since been largely replaced by Delta, and the current third wave (began in December 2021) is dominated by Delta and the highly transmissible Omicron variant.
We performed a subanalysis of SARS-CoV-2 variants during the third wave (Figure 2, panel D). We also detected other Delta sublineages, such as AY.1 or B.1.617.2.1 (also known as Delta Plus and associated with a relatively higher transmissibility) (6), at a low prevalence. The AY.1 Delta sublineage has been associated with more antibody escaping properties because of the K417N mutation, which was identified in the Beta variant (7). We also provide the relative number of mutations for SARS-CoV-2 variants (Figure 2, panel E). We deposited all sequences generated during this study in the GISAID public database (accession nos. EPI ISL 4548461–543, EPI_ISL_6262724–47, EPI_ISL_8307285–411, EPI_ISL_8523904–5, EPI_ISL_6506618, EPI_ISL_6506627, EPI_ISL_6506639, EPI_ISL_6506648, EPI_ISL_6506655, EPI_ISL_6506666, EPI_ISL_6506674, EPI_ISL_6506689, EPI_ISL_6506697, EPI_ISL_6506706, EPI_ISL_6506713, EPI_ISL_6506721, EPI_ISL_6506726, EPI_ISL_6506738, EPI_ISL_6506747, EPI_ISL_6506751, EPI_ISL_6506760, EPI_ISL_6506767, EPI_ISL_6506773, EPI_ISL_6506784, EPI_ISL_6506791, EPI_ISL_6506802, EPI_ISL_6506812, EPI_ISL_6506824, EPI_ISL_6506829, EPI_ISL_6506835, EPI_ISL_6506841, EPI_ISL_6506844, EPI_ISL_6506851, and EPI_ISL_6506857).
SARS-CoV-2 sequences deposited in GISAID from Uganda showed a rapid replacement of variants since the beginning of the COVID-19 pandemic. Genomic sequencing involving 266 samples collected during June‒December 2021 showed that the Delta variant was the dominant virus. However, the Omicron variant emerged in late November 2021 from travelers arriving through Entebbe International Airport (39.29% from South Africa, 28.57% from Nigeria, 14.29% from Kenya, 7.14% from the Democratic Republic of the Congo, 3.57% from Ethiopia, 3.57% Rwanda, and 3.57% from the United States), and Omicron community transmissions are increasing (based on PCR genotyping). Therefore, we anticipate that Delta is gradually being replaced by Omicron, which is consistent with the observed SARS-CoV-2 variants trajectory over time.
Furthermore, results from a mutation-specific SARS-CoV-2 PCR screening (8,9) suggest that Omicron, initially becoming dominant among travelers, will likely later predominate in the community. The Omicron variant has been associated with increased transmissibility and has quickly become a global concern (10). Speeding up genomic sequencing from prospective samples collected at points of entry and from the community will enable faster response to outbreaks as they emerge.
A major limitation of this study was suboptimal sampling. Previously, convenience sampling that targeted points of entry and outbreak hotspots was more common. Sampling prioritized mostly moderate-to-high community transmission sites and focused less on sampling low viral transmission communities. However, plans are under way to adopt effective sampling guidelines to ensure geographically representative sampling (11,12).
In summary, the SARS-CoV-2 Delta variant rapidly replaced earlier virus variants after it was introduced into Uganda. The Omicron variant has followed the same trajectory. Our results highlight the need for surveillance and infection control measures to prevent future pandemic waves.
Dr. Bbosa is a postdoctoral scientist at the Medical Research Council/Uganda Virus Research Institute and the London School of Hygiene and Tropical Medicine Uganda Research Unit, Entebbe, Uganda. His primary research interests are viral genomics, molecular epidemiology, pathogen phylodynamics and bioinformatics, and infectious disease epidemic response.
Acknowledgments
We thank the Uganda Ministry of Health and its COVID-19 Scientific Advisory Committee, the National COVID-19 Task Force, and the staff of the Emerging and Remerging Infections Department of the Uganda Virus Research Institute for their providing contributions; the team at the KwaZulu-Natal Research Innovation and Sequencing Platform for providing laboratory training and support; the staff at the Uganda Virus Research Institute for collecting field samples; and the persons who collected samples at borders or points of entry into Uganda.
This study was supported by the United Kingdom Medical Research Council and the United Kingdom Department for International Development under their concordat agreement. Training in genomic sequencing was supported by the African Society of Laboratory Medicine and Africa Centres for Disease Control and Prevention.
References
- Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–60.PubMedGoogle Scholar
- Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, et al. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int J Surg. 2020;78:185–93. DOIPubMedGoogle Scholar
- Bugembe DL, Kayiwa J, Phan MVT, Tushabe P, Balinandi S, Dhaala B, et al. Main routes of entry and genomic diversity of SARS-CoV-2, Uganda. Emerg Infect Dis. 2020;26:2411–5. DOIPubMedGoogle Scholar
- Bugembe DL, Phan MVT, Ssewanyana I, Semanda P, Nansumba H, Dhaala B, et al. Emergence and spread of a SARS-CoV-2 lineage A variant (A.23.1) with altered spike protein in Uganda. Nat Microbiol. 2021;6:1094–101. DOIPubMedGoogle Scholar
- Vilsker M, Moosa Y, Nooij S, Fonseca V, Ghysens Y, Dumon K, et al. Genome Detective: an automated system for virus identification from high-throughput sequencing data. Bioinformatics. 2019;35:871–3. DOIPubMedGoogle Scholar
- Kannan SR, Spratt AN, Cohen AR, Naqvi SH, Chand HS, Quinn TP, et al. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J Autoimmun. 2021;124:
102715 . DOIPubMedGoogle Scholar - Rahman FI, Ether SA, Islam MR. The “Delta Plus” COVID-19 variant has evolved to become the next potential variant of concern: mutation history and measures of prevention. J Basic Clin Physiol Pharmacol. 2021;33:109–12. DOIPubMedGoogle Scholar
- Primerdesign Ltd. SNPsig® real-time PCR SARS-CoV-2 mutation detection/allelic discrimination kit. Identification of L452R mutation [cited 2022 Jan 13]. https://genesig.com/assets/files/snpsig_sars_cov_2_l452r_version_1_01.pdf
- Wang H, Jean S, Eltringham R, Madison J, Snyder P, Tu H, et al. Mutation-specific SARS-CoV-2 PCR screen: rapid and accurate detection of variants of concern and the identification of a newly emerging variant with spike L452R mutation. J Clin Microbiol. 2021;59:
e0092621 . DOIPubMedGoogle Scholar - Thakur V, Ratho RK. OMICRON (B.1.1.529): a new SARS-CoV-2 variant of concern mounting worldwide fear. J Med Virol. 2021; [Epub ahead of print].PubMedGoogle Scholar
- Ministry of Health Uganda. A guide for genomic surveillance of COVID-19 in Uganda. The Ministry: Kampala (Uganda); 2021.
- Africa CDC and WHO. Interim operational guidance on SARS-CoV-2 genomic surveillance in Africa: an updated guide. 2021 [cited 2022 Jan 10]. https://africacdc.org/download/interim-operational-guidance-on-sars-cov-2-genomic-surveillance-in-africa-an-updated-guide
Figures
Cite This ArticleOriginal Publication Date: March 23, 2022
Table of Contents – Volume 28, Number 5—May 2022
| EID Search Options |
|---|
|
|
|
|
|
|
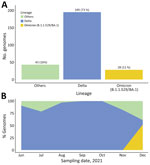
![Rapid replacement of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants by Delta and subsequent arrival of Omicron, Uganda, 2021. A) Coronavirus disease pandemic waves. Confirmed cases of daily coronavirus disease and trends of the SARS-CoV-2 pandemic over time. Three waves of the pandemic dominated by the A.23.1 in the first wave (December 2020‒January 2021), Delta in the second wave (May‒July 2021), and the Delta and Omicron variants in the third wave (Omicron emerged in late November 2021 and the wave began in December 2021). B, C) SARS-CoV-2 variants over time (950 genomes deposited in the GISAID database [https://www.gisaid.org] by January 10, 2022). D) SARS-CoV-2 variants during the third wave among travelers and community samples. E) Violin plots showing the distribution of whole-genome nucleotide mutations in each of the SARS-CoV-2 lineages by using the wild-type Wuhan-Hu-1/2019 isolate (GenBank accession no. MN908947) as the reference. Black dots indicate median number of nucleotide mutations. Error bars indicate interquartile ranges.](/eid/images/22-0121-F2-tn.jpg)
Please use the form below to submit correspondence to the authors or contact them at the following address:
Nicholas Bbosa, Medical Research Council/Uganda Virus Research Institute and London School of Hygiene and Tropical Medicine Uganda Research Unit, Plot 51‒59 Nakiwogo Rd, Entebbe, Uganda
Top