Outbreak of Imported Seventh Pandemic Vibrio cholerae O1 El Tor, Algeria, 2018
Nabila Benamrouche

, Chafika Belkader, Elisabeth Njamkepo, Sarah Sihem Zemam, Soraya Sadat, Karima Saighi, Dalila Torkia Boutabba, Faiza Mechouet, Rym Benhadj-Slimani, Fatma-Zohra Zmit, Jean Rauzier, Farid Kias, Souad Zouagui, Corinne Ruckly, Mohamed Yousfi, Amel Zertal, Ramdane Chouikrat, Marie-Laure Quilici, and François-Xavier Weill

Author affiliations: Institut Pasteur d’Algérie, Algiers, Algeria (N. Benamrouche, C. Belkader, S.S. Zemam, S. Sadat, D.T. Boutabba, R. Benhadj-Slimani, F. Kias); University of Algiers I, Algiers (N. Benamrouche, F. Mechouet, F.-Z. Zmit, A. Zertal); Institut Pasteur, Université Paris Cité, Paris, France (E. Njamkepo, J. Rauzier, C. Ruckly, M.-L. Quilici, F.-X. Weill); Public Hospital Establishment of Boufarik, Blida, Algeria (K. Saighi, M. Yousfi, R. Chouikrat); Specialized Hospital Establishment El Hadi Flici, Algiers (F. Mechouet, F.-Z. Zmit, A. Zertal); University Hospital of Oran and University of Oran I, Oran, Algeria (S. Zouagui)
Main Article
Figure 2
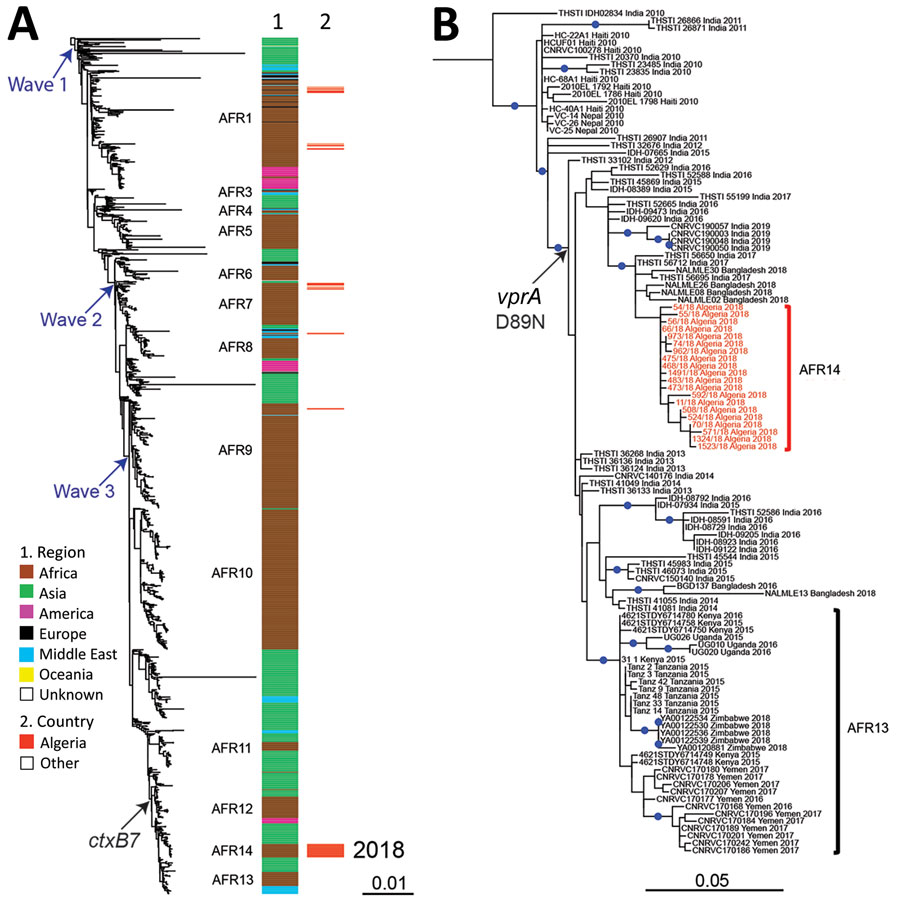
Figure 2. Phylogenomic analyses of Vibrio cholerae O1 El Tor isolates from Algeria, 2018. A) Maximum-likelihood phylogeny for 1,285 seventh pandemic V. cholerae biotype El Tor genomic sequences. A6 was used as the outgroup (Appendix 1, Table 4). Genomic waves and acquisition of ctxB7 allele are indicated. Sublineages previously introduced into Africa (AFR1, AFR3–AFR13) are shown at the right of the tree. Column 1 indicates the geographic origins of the isolates; column 2 indicates isolates from the 2018 cholera outbreak in Algeria, all of which belong to a new seventh pandemic wave 3 sublineage AFR14. B) Maximum-likelihood phylogeny for 115 wave 3 ctxB7 isolates belonging to the distal part of the tree in panel A. N16961 was used as the outgroup (Appendix 1, Table 4). The isolates belonging to AFR14 from the 2018 cholera outbreak in Algeria are shown in red. Acquisition of the polymyxin susceptibility–associated single nucleotide variant in vprA (D89N) is indicated. Blue dots indicate bootstrap values >90%. Scale bars indicate the number of nucleotide substitutions per variable site.
Main Article
Page created: March 23, 2022
Page updated: May 22, 2022
Page reviewed: May 22, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
