Volume 28, Number 7—July 2022
Dispatch
Chronic Pulmonary Disease Caused by Tsukamurella toyonakaense
Cite This Article
Citation for Media
Abstract
Unidentified Mycobacterium species are sometimes detected in respiratory specimens. We identified a novel Tsukamurella species (Tsukamurella sp. TY48, RIMD 2001001, CIP 111916T), Tsukamurella toyonakaense, from a patient given a misdiagnosis of nontuberculous mycobacterial pulmonary disease caused by unidentified mycobacteria. Genomic identification of this Tsukamurella species helped clarify its clinical characteristics and epidemiology.
In clinical practice, unidentified Mycobacterium species are sometimes detected in respiratory specimens. Few Mycobacterium species can be identified by using methods available in clinical practice, although there are ≈200 species of nontuberculous mycobacteria (NTM) (1). We reported a case of pulmonary disease caused by a novel mycobacteria species identified by using multilocus sequence typing (MLST) and whole-genome sequencing (WGS) (2).
We investigated the epidemiology of unidentified pathogenic mycobacteria by using TRCReady MTB and MAC (Tosoh Bioscience, https://www.tosohbioscience.com), AccuProbe (Gen-Probe Inc., https://www.gen-probe.com), COBAS AMPLICOR (Roche Diagnostics, https://www.roche.com), and a DNA–DNA hybridization assay (Kyokuto Pharmaceutical Industrial, https://www.kyokutoseiyaku.co.jp). WGS analysis of preserved unidentified mycobacteria culture isolates was approved by the institutional research ethics board (TNH2019063–2). The requirement for informed consent was waived because of the retrospective nature of the analysis. The opt-out recruitment method was applied to provide an opportunity for all patients to decline participation. Results of WGS analysis of TY48 were deposited in BioProject (accession no. PRJDB10620) and BioSample (accession no. SAMD00250050).
We performed MLST and WGS of culture isolates from 8 patients given diagnoses of NTM pulmonary disease caused by unidentified mycobacteria. We identified Mycobacterium shimoidei, M. shinjukuense, M. paragordonae, M. heckeshornense, M. lentiflavum (3 isolates), and a novel Tsukamurella species (Tsukamurella sp. TY48, RIMD 2001001, CIP 111916T).
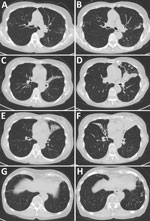
The patient with Tsukamurella infection was an 82-year-old woman who had received a diagnosis of NTM pulmonary disease 23 years earlier. Then a 59-year-old previously healthy woman, she was referred to our hospital because of abnormal chest radiographic findings. Although she had no symptoms, chest computed tomography findings showed centrilobular nodules and bronchiectasis. During follow-up, a cough and occasional hemoptysis developed. M. chelonae was repeatedly identified from her sputum. We started airway clearance therapy with erythromycin and expectorants. After 2 years of treatment, the M. chelonae disappeared from her sputum. However, her symptoms and radiologic findings slowly but steadily progressed (Figure 1), and rapidly growing acid-fast bacilli were repeatedly detected in her sputum for 8 years. The culture isolates were Ziehl-Neelsen stain positive. However, the species/subspecies could not be identified by using conventional methods. Therefore, she was given a diagnosis of NTM pulmonary disease caused by unidentified mycobacteria.
We continued erythromycin treatment for >20 years on the basis of evidence regarding successful treatment of NTM pulmonary disease with erythromycin (3). However, her symptoms and radiologic findings of lung destruction and structural alterations slowly but steadily progressed.
Because of this progression, we performed WGS by using a MinION Sequencer and Flow Cell R94 (Oxford Nanopore Technologies, https://nanoporetech.com). We extracted genomic DNA from cultured isolates by using a NucleoSpin Microbial DNA Kit (Takara Bio, https://www.takarabio.com) and prepared a library by using the Rapid Barcoding Kit (Oxford Nanopore Technologies). Using MinION raw sequencing reads, we performed MLST analysis on the 184-gene accessory genome with mlstverse software (https://www.multiverse.io) as reported (1). The unidentified mycobacterium was presumed to be M. fallax (MLST score 0.083). However, the low MLST score prompted a deeper analysis of the bacterial genome.
We conducted a 16S rRNA analysis by performing a homology search using blastn (https://blast.ncbi.nlm.nih.gov) and compared our data with that in the SILVA rRNA database (4). The phylogenetic tree constructed using full-length 16S rRNA genes showed that strain TY48 was closely related to other Tsukamurella species (>98.7%), whereas its homology to 2 type species belonging to the related bacteria Gordonia bronchialis and Williamsia muralis was only 94.0% (Figure 2). We next determined the complete genome sequence of TY48 as reported (1) and performed WGS by using MinION and HiSeq 2500 instruments (Illumina, https://www.illumina.com). We performed genome assembly for strain TY48 by using flye (https://www.flye.com) for long reads obtained from MinION and corrected sequencing error by using pilon (https://bio.tools/pilon).
A comparison of the TY48 genome sequence with those of other Tsukamurella species indicated that the nearest related species was T. paurometabola (average nucleotide identity of 86.2%) (Table 1). This finding suggested that Tsukamurella sp. TY48 (RIMD 2001001; CIP 111916T) was a novel Tsukamurella species.
We performed antimicrobial drug susceptibility tests for rapidly growing mycobacteria by using the broth microdilution method in accordance with Clinical and Laboratory Standards Institute M24-A2 guidelines (5). We transferred the culture to Middlebrook 7H9 broth and vortexed. We adjusted the culture medium to a 0.5 McFarland standard with sterile distilled water; we then added 60 μL of the 0.5 McFarland suspension to a Cation-Adjusted Mueller-Hinton Broth (Kyokuto Pharmaceutical Industrial Co. Ltd., https://www.kyokutoseiyaku.co.jp) and dispensed 100 μL of this solution into each well of the panel. After confirming adequate growth of the control over a 3-day incubation in a standard atmosphere at 30°C, we determined the MICs (μg/mL) for 15 drugs: clarithromycin, 0.25; azithromycin, <0.25; cefoxitin, <8; imipenem, <0.5; meropenem, <0.5; faropenem, <1; amikacin, <1; tobramycin, 2; minocycline, <0.25; doxycycline, <1; linezolid, <4; moxifloxacin, <0.25; ciprofloxacin <0.5; levofloxacin, <0.5; and trimethoprim/sulfamethoxazole, <2/38. Tsukamurella sp. TY48 was sensitive to all 15 drugs.
We renamed TY48 as T. toyonakaense after the location of its discovery, Toyonaka, Japan. T. toyonakaense is an aerobic, nonmotile, gram-positive rod that grows at 30°C and 37°C, but not at 42°C, and produces catalase. After a 72-h incubation at 30°C on 7H11 agar, it forms white and creamy, rough, nonpigmented colonies (10 mm in diameter). According to the API 50 CH system (bioMérieux, https://www.biomerieux.com), this bacterium can assimilate fructose, glucose, starch, sucrose, and trehalose but not arabinose, mannitol, mannose, or xylose.
After diagnosis, we attempted combination drug therapy with clarithromycin (200 mg/d) and ethambutol (250 mg/d). The patient refused continuation of treatment after 2 weeks because of antimicrobial drug–induced fatigue. We then resumed treatment with erythromycin. Her symptoms and radiologic findings are slowly improving (Table 2; Appendix).
Tsukamurella species are aerobic, gram-positive, partially acid-fast, and nonmotile bacilli that can cause opportunistic infections, including pulmonary disease (6). Sixteen species of Tsukamurella have been classified (7). Only 9 pulmonary disease cases have been reported (8,9) (Table 2).
The prevalence of Tsukamurella pulmonary disease is probably underestimated. The genus Tsukamurella is often misidentified as related genera because it is difficult to identify in most clinical microbiology laboratories (10). Because of its partially acid-fast bacilli and cavitary shadow in radiologic examination, Tsukamurella pulmonary disease is often confused with Mycobacterium infection and often treated with antituberculous drugs (9). Yu et al. genotyped specimens from 101 NTM pulmonary disease patients by using 16S rRNA and 16S‒23S rRNA internal transcribed spacer sequences and detected Tsukamurella species in ≈1% of the specimens (11). If one considers the prevalence of NTM pulmonary disease, the actual prevalence of Tsukamurella pulmonary disease is probably much higher than the 9 reported cases.
Tsukamurella commonly causes acute onset pneumonia with cavity and consolidation (Table 2) and fever, coughing, sputum, fatigue, and hemoptysis. Although appropriate drugs and treatment durations are unknown, combination medications of >2 drugs, including rifampin or quinolone, are widely used and presumed effective on the basis of case reports (6,8,12–14). These reports also indicated a good prognosis for Tsukamurella pulmonary disease (8,12,13). No relapses were reported, in contrast to NTM pulmonary disease. Although the Clinical and Laboratory Standards Institute has proposed breakpoints for aerobic actinomycetes (5), no definitive drug breakpoints for Tsukamurella spp. have been established. However, the strain we identified showed extensive antimicrobial drug susceptibility.
Because a clinically applicable identification technique is not available, Tsukamurella infections are probably underestimated and more prevalent than has been recognized. Misidentification as related genera, especially Mycobacterium, results in missed opportunities to properly treat Tsukamurella infections. Use of genomic sequencing to identify Tsukamurella species and more cases of Tsukamurella infections will help identify clinical characteristics and clarify epidemiology of Tsukamurella pulmonary disease.
Dr. Kuge is a clinical fellow in respiratory medicine in the respiratory medicine department of Osaka University Medical Hospital, Osaka, Japan. His primary research interest is respiratory infections.
Acknowledgments
We thank the patient for granting permission to publish her information and Traci Raley and Katie Oakley for editing drafts of this manuscript.
This study was supported in part by the Japan Agency for Medical Research and Development (grants JP20fk0108129, JP21fk0108129h0702, and JP21lm0203007), GlaxoSmithKline (grant A-32), the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (grants JP21K16118 and JP21K08194), the Smoking Research Foundation, the Takeda Science Foundation, and the Japan Intractable Diseases (Nanbyo) Research Foundation (grant 2020B02).
K.F. designed the study; K.H., H.S., T.N., and A.K. performed mycobacterial culture and analysis of culture isolates; K.F. and T.K. performed clinical and laboratory data extraction and analysis; Y.A., T.S., E.A., Takahiro Kawasaki, T.M., K.T., M.M., K.M., and H.K. assisted with data extraction and analysis; Y.M., D.M., and S.N. performed multilocus typing analysis and whole-genome analysis; M.T., Y.H., and T.I. analyzed biochemical profiles of the strain; K.F. and S.K. wrote the manuscript; and A.K. and H.K. supervised the study. All authors read and approved the manuscript.
References
- Matsumoto Y, Kinjo T, Motooka D, Nabeya D, Jung N, Uechi K, et al. Comprehensive subspecies identification of 175 nontuberculous mycobacteria species based on 7547 genomic profiles. Emerg Microbes Infect. 2019;8:1043–53. DOIPubMedGoogle Scholar
- Kuge T, Fukushima K, Matsumoto Y, Abe Y, Akiba E, Haduki K, et al. Pulmonary disease caused by a newly identified mycobacterium: Mycolicibacterium toneyamachuris: a case report. BMC Infect Dis. 2020;20:888. DOIPubMedGoogle Scholar
- Komiya K, Kurashima A, Ihi T, Nagai H, Matsumoto N, Mizunoe S, et al. Long-term, low-dose erythromycin monotherapy for Mycobacterium avium complex lung disease: a propensity score analysis. Int J Antimicrob Agents. 2014;44:131–5. DOIPubMedGoogle Scholar
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):D590–6. DOIPubMedGoogle Scholar
- Woods GL, Brown-Elliott BA, Conville PS, Desmond EP, Hall GS, Lin G, et al. Susceptibility testing of Mycobacteria, Nocardiae, and other aerobic Actinomycetes. 2nd ed. Wayne (PA): Clinical and Laboratory Standards Institute; 2011.
- Mehta YB, Goswami R, Bhanot N, Mehta Z, Simonelli P. Tsukamurella infection: a rare cause of community-acquired pneumonia. Am J Med Sci. 2011;341:500–3. DOIPubMedGoogle Scholar
- Teng JLL, Fong JYH, Fok KMN, Lee HH, Chiu TH, Tang Y, et al. Tsukamurella asaccharolytica sp. nov., Tsukamurella conjunctivitidis sp. nov. and Tsukamurella sputi sp. nov., isolated from patients with bacteraemia, conjunctivitis and respiratory infection in Hong Kong. Int J Syst Evol Microbiol. 2020;70:995–1006. DOIPubMedGoogle Scholar
- Inchingolo R, Nardi I, Chiappini F, Macis G, Ardito F, Sali M, et al. First case of Tsukamurella pulmonis infection in an immunocompetent patient. Respir Med CME. 2010;3:23–5. DOIGoogle Scholar
- Yang L, Cao Y, Dan Z, Wang Z, Wang X. Community-acquired Tsukamurella pneumonia in a young immunocompetent adult: a case misdiagnosed as pulmonary tuberculosis and literature review. Postgrad Med. 2017;129:563–6. DOIPubMedGoogle Scholar
- Teng JLL, Tang Y, Wong SSY, Fong JYH, Zhao Z, Wong CP, et al. MALDI-TOF MS for identification of Tsukamurella species: Tsukamurella tyrosinosolvens as the predominant species associated with ocular infections. Emerg Microbes Infect. 2018;7:80. DOIPubMedGoogle Scholar
- Yu XL, Lu L, Chen GZ, Liu ZG, Lei H, Song YZ, et al. Identification and characterization of non-tuberculous mycobacteria isolated from tuberculosis suspects in Southern-central China. PLoS One. 2014;9:
e114353 . DOIPubMedGoogle Scholar - Alcaide ML, Espinoza L, Abbo L. Cavitary pneumonia secondary to Tsukamurella in an AIDS patient. First case and a review of the literature. J Infect. 2004;49:17–9. DOIPubMedGoogle Scholar
- Perez VA, Swigris J, Ruoss SJ. Coexistence of primary adenocarcinoma of the lung and Tsukamurella infection: a case report and review of the literature. J Med Case Reports. 2008;2:207. DOIPubMedGoogle Scholar
- Maalouf R, Mierau SB, Moore TA, Kaul A. First case report of community-acquired pneumonia due to Tsukamurella pulmonis. Ann Intern Med. 2009;150:147–8. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 28, Number 7—July 2022
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Kiyoharu Fukushima, Department of Respiratory Medicine and Clinical Immunology, Osaka University Graduate School of Medicine, 2-2 Yamadaoka, Suita, Osaka, Japan; e-mail:fukushima@imed3.med.osaka-u.ac.jp
Top