Volume 28, Number 8—August 2022
Research Letter
Lymphocytic Choriomeningitis Virus Infection, Australia
Figure
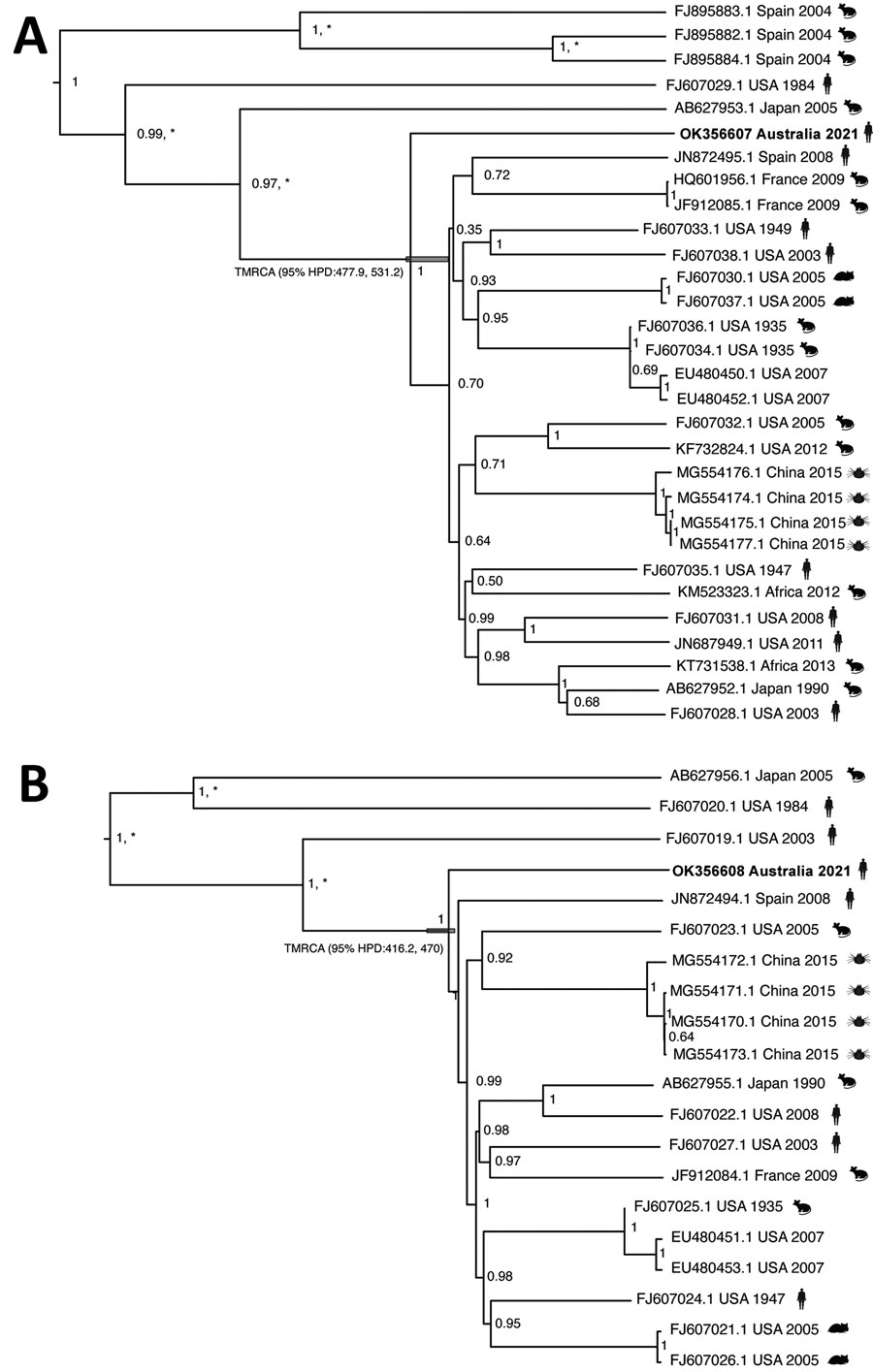
Figure. Phylogenetic relationships of a strain of lymphocytic choriomeningitis virus from a man in Australia and the broader lymphocytic choriomeningitis virus phylogeny. Tips are labeled with GenBank sequence accession number, country of origin, year of collection, and host (mice, hamsters, humans, ticks). Trees were generated by using BEAST 1.10.4 (9) to estimate the time to the most recent common ancestor between the novel virus sequence and its closest phylogenetic relative. We used the Hasegawa-Kishino-Yano plus gamma substitution model with a strict clock and an exponential growth coalescent tree prior. Because the dataset exhibits high sequence divergence, we calibrated the molecular clock by using previous independent estimates of the substitution rate, with a fixed clock rate for the long segment of 3.7 × 10–4 substitutions/site/year and 3.3 × 10–4 substitutions/site/year for the short segment (10). Highest clade credibility tree of the short segment (GenBank accession no. OK356607) sequences (n = 29) (A) and highest clade credibility tree of the long segment (GenBank accession no. OK356608) sequences (n = 19) (B). Node labels denote the posterior support, and an asterisk represents a bootstrap percentage of >70% support for a specific clade, using 1,000 ultra-fast bootstrap replicates in a maximum-likelihood tree approach using IQ-TREE2 (11). The 95% highest posterior density for the divergence time before present of the Australia sample is annotated in the respective node.
References
- Armstrong C, Lillie RD. Experimental lymphocytic choriomeningitis of monkeys and mice produced by a virus encountered in studies of the 1933 St. Louis encephalitis epidemic. Public Health Reports (1896–1970). 1934;49:1019–27.
- Lewis JM, Utz JP. Orchitis, parotitis and meningoencephalitis due to lymphocytic-choriomeningitis virus. N Engl J Med. 1961;265:776–80. DOIPubMedGoogle Scholar
- Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, et al. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–8. DOIPubMedGoogle Scholar
- Bonthius DJ, Wright R, Tseng B, Barton L, Marco E, Karacay B, et al. Congenital lymphocytic choriomeningitis virus infection: spectrum of disease. Ann Neurol. 2007;62:347–55. DOIPubMedGoogle Scholar
- Gregg MB. Recent outbreaks of lymphocytic choriomeningitis in the United States of America. Bull World Health Organ. 1975;52:549–53.PubMedGoogle Scholar
- Holdsworth RL, Downie E, Georgiades MJ, Bradbury R, Druce J, Collett J. Lymphocytic choriomeningitis virus in western New South Wales. Med J Aust. 2022;216:71–2. DOIPubMedGoogle Scholar
- Kafetzopoulou LE, Efthymiadis K, Lewandowski K, Crook A, Carter D, Osborne J, et al. Assessment of metagenomic Nanopore and Illumina sequencing for recovering whole genome sequences of chikungunya and dengue viruses directly from clinical samples. Euro Surveill. 2018;23:23. DOIPubMedGoogle Scholar
- Gabriel SI, Stevens MI, Mathias ML, Searle JB. Of mice and ‘convicts’: origin of the Australian house mouse, Mus musculus. PLoS One. 2011;6:
e28622 . DOIPubMedGoogle Scholar - Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:
vey016 . DOIPubMedGoogle Scholar - Albariño CG, Palacios G, Khristova ML, Erickson BR, Carroll SA, Comer JA, et al. High diversity and ancient common ancestry of lymphocytic choriomeningitis virus. Emerg Infect Dis. 2010;16:1093–100. DOIPubMedGoogle Scholar
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar