Volume 28, Number 9—September 2022
Dispatch
Laboratory Misidentifications Resulting from Taxonomic Changes to Bacillus cereus Group Species, 2018–2022
Figure 1
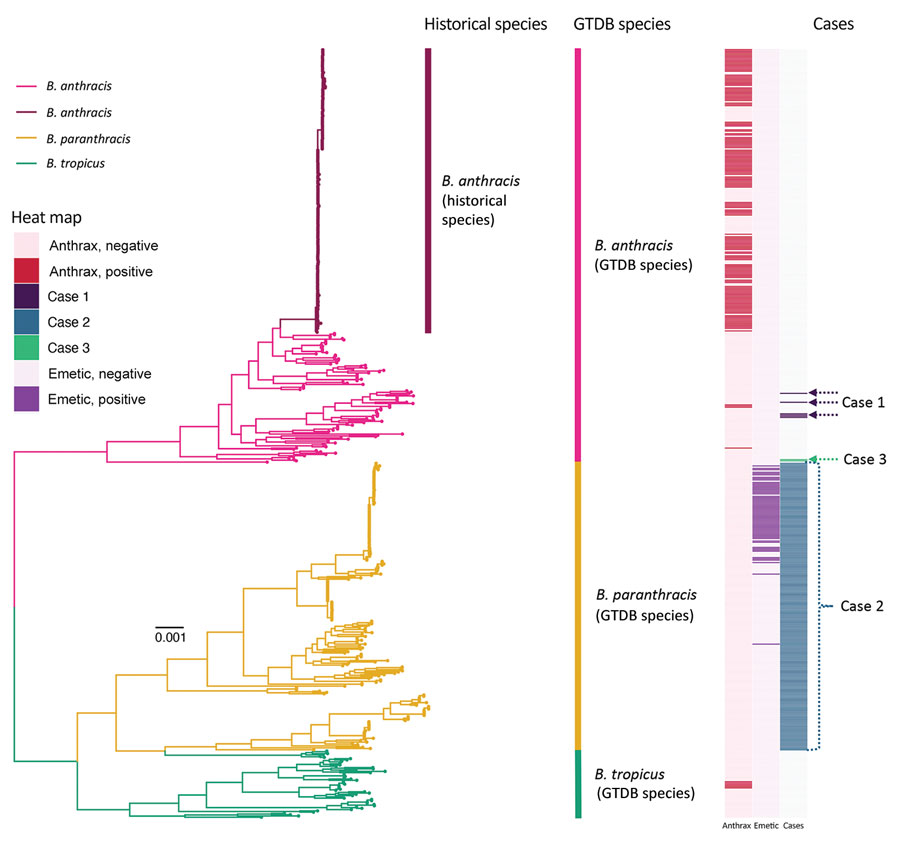
Figure 1. Maximum-likelihood phylogeny constructed using core genes detected among 605 genomes assigned to the GTDB B. anthracis, B. paranthracis, and B. tropicus species. Branch colors and clade labels denote GTDB species assignments or, for B. anthracis, historical species assignments. The heatmap to the right of the phylogeny shows whether a strain possessed anthrax toxin-encoding genes or not (anthrax); whether a strain possessed cereulide synthetase (emetic toxin)-encoding genes or not (emetic); and the strains or lineages discussed in the cases we detailed here (cases) (Table 1). For case 1, the actual genomes were not publicly available; thus, genomes assigned to the same sequence types (STs, via 7-gene multilocus sequence typing) are highlighted. For case 2, the only information provided to the authors was that the genome in question belonged to species B. paranthracis; thus, all genomes assigned to GTDB’s B. paranthracis species are highlighted. For case 3, the actual strain genomes associated with the case are highlighted. The phylogeny was rooted using panC Group II B. cereus group strain FSL W8-0169 as an outgroup (National Center for Biotechnology Information RefSeq Assembly accession no. GCF_001583695.1; omitted for readability). Branch lengths are reported in substitutions per site. GTDB, Genome Taxonomy Database.