Volume 29, Number 1—January 2023
Dispatch
Survey of West Nile and Banzi Viruses in Mosquitoes, South Africa, 2011–2018
Figure 2
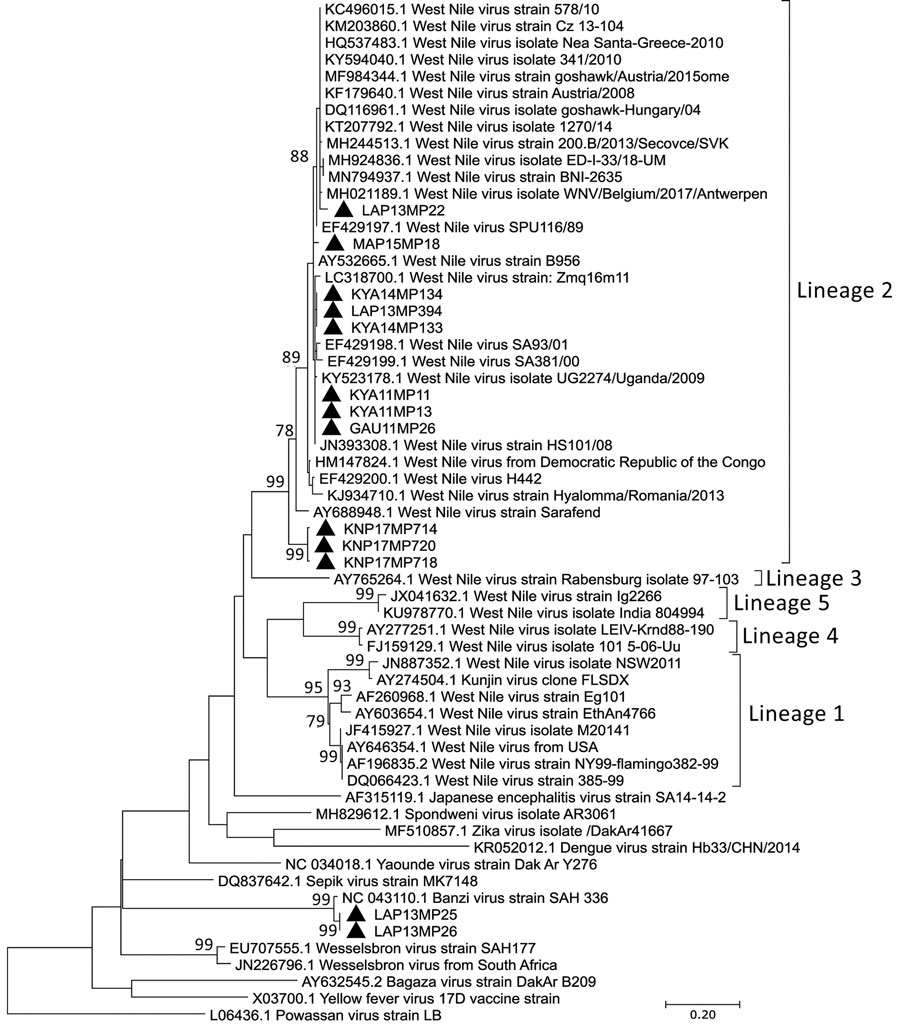
Figure 2. Phylogenetic analysis of flaviviruses using NS5 gene sequences in survey of West Nile and Banzi viruses in mosquitoes, South Africa, 2011–2018. Maximum likelihood analysis was used to identify flaviviruses found in mosquitoes after partial sequencing of the flavivirus NS5 gene region (226 nt, Kimura 2-parameter model plus gamma distribution plus proportion of invariable sites). Sequence data were edited by using CLC Main Workbench version 8.0.1 (QIAGEN, https://www.qiagen.com). Reference genomes were downloaded from GenBank. Multiple sequence alignments were created by using MAFFT (https://mafft.cbrc.jp/alignment/server/index.html) with default parameters. Phylogenetic analysis was performed by using MEGA X software (MEGA, https://www.megasoftware.net) with bootstrap support for network groupings calculated from 1,000 replicates. Bootstrap values (>70%) are displayed on branches. GenBank accession numbers for newly sequenced virus strains: OL411950 (KYA11MP13 isolate), OL411951 (GAU11MP26 isolate), OL411952 (KYA14MP133 isolate), OL411953 (KYA14MP134 isolate), OL411954 (LAP14MP394 isolate), OL411955 (MAR15MP18 isolate), OL411956 (LAP13MP22 isolate), OL411957 (KNP17MP714 isolate), OL411958 (KNP17MP720 isolate), OL411959 (KNP17MP718 isolate), OL411960 (KYA11MP11 isolate), OL411961 (LAP13MP25 isolate), and OL411962 (LAP13MP26 isolate). Solid black triangles are new viral sequences that were detected in mosquitoes in this study. Scale bar indicates nucleotide substitutions per site.