Volume 29, Number 10—October 2023
Research
Candida auris Clinical Isolates Associated with Outbreak in Neonatal Unit of Tertiary Academic Hospital, South Africa
Figure 2
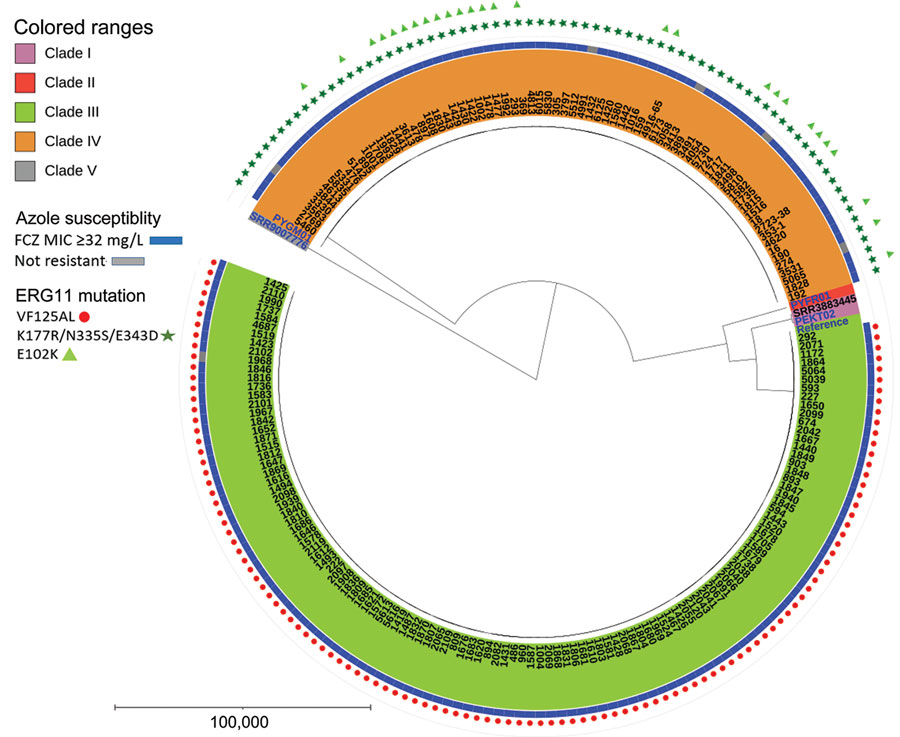
Figure 2. Phylogenetic tree depicting clade distribution and fluconazole resistance mutations of 188 invasive or colonizing South African Candida auris strains isolated from patients admitted to a large metropolitan hospital in South Africa, 2016–2020. The unrooted maximum-parsimony tree was created using MEGA software (https://www.megasoftware.net) using 287,338 single-nucleotide polymorphisms based on 1,000 bootstrap replicates. Scale bar indicates number of pairwise differences. FCZ, fluconazole.
1Members of GERMS-SA are listed at the end of this article.
Page created: August 31, 2023
Page updated: September 20, 2023
Page reviewed: September 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.