Volume 29, Number 10—October 2023
Research
Ancestral Origin and Dissemination Dynamics of Reemerging Toxigenic Vibrio cholerae, Haiti
Figure 4
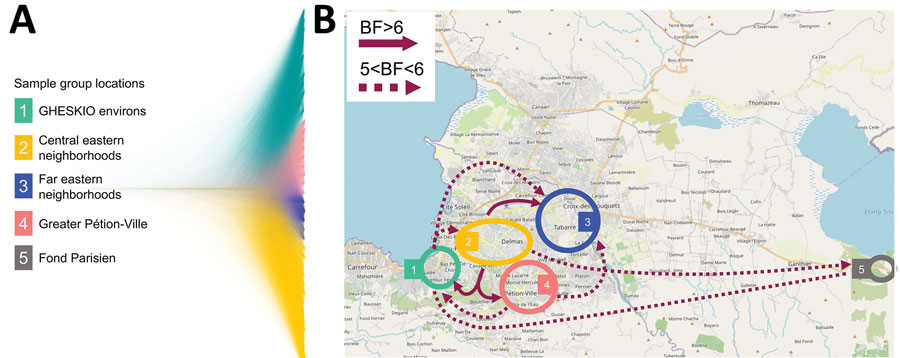
Figure 4. Bayesian phylogeography and dissemination patterns of reemerging toxigenic Vibrio cholerae, Haiti, during the 2022 outbreak. A) Bayesian DensiTree showing superimposed posterior distribution of trees inferred by the phylogeography analysis of V. cholerae full genomes from clinical cases sampled in Haiti during October 3–November 21. Colors indicate branches of samples grouped by location. B) Locations of case clusters delimited by colored circles. Solid- and broken-lined arrows indicate migration patterns among areas, as supported by Bayes factor and inferred from phylogeographic analysis by using a discrete trait asymmetric diffusion model. We considered rates yielding a BF >3 supported diffusion rates (33) and BF >6 decisive support (34), constituting the migration graph. Maps demonstrate major migrations of 5<BF<6 and BF >6. A list of all migrations supported by BF >3 are reported elsewhere (Appendix Table 5). BF, Bayes factor.
References
- World Health Organization. Cholera—global situation. 2022 [cited 2023 Feb 8]. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON426
- Hendriksen RS, Price LB, Schupp JM, Gillece JD, Kaas RS, Engelthaler DM, et al. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. MBio. 2011;2:e00157–11. DOIPubMedGoogle Scholar
- Piarroux R, Barrais R, Faucher B, Haus R, Piarroux M, Gaudart J, et al. Understanding the cholera epidemic, Haiti. Emerg Infect Dis. 2011;17:1161–8. DOIPubMedGoogle Scholar
- Ivers LC, Walton DA. The “first” case of cholera in Haiti: lessons for global health. Am J Trop Med Hyg. 2012;86:36–8. DOIPubMedGoogle Scholar
- Blackburn JK, Diamond U, Kracalik IT, Widmer J, Brown W, Morrissey BD, et al. Household-level spatiotemporal patterns of incidence of cholera, Haiti, 2011. Emerg Infect Dis. 2014;20:1516–9. DOIPubMedGoogle Scholar
- Alam MT, Weppelmann TA, Longini I, De Rochars VM, Morris JG Jr, Ali A. Increased isolation frequency of toxigenic Vibrio cholerae O1 from environmental monitoring sites in Haiti. PLoS One. 2015;10:
e0124098 . DOIPubMedGoogle Scholar - Alam MT, Weppelmann TA, Weber CD, Johnson JA, Rashid MH, Birch CS, et al. Monitoring water sources for environmental reservoirs of toxigenic Vibrio cholerae O1, Haiti. Emerg Infect Dis. 2014;20:356–63. DOIPubMedGoogle Scholar
- Mavian C, Paisie TK, Alam MT, Browne C, Beau De Rochars VM, Nembrini S, et al. Toxigenic Vibrio cholerae evolution and establishment of reservoirs in aquatic ecosystems. Proc Natl Acad Sci U S A. 2020;117:7897–904. DOIPubMedGoogle Scholar
- World Health Organization. Cholera—Haiti, 12 October 2022 [cited 2023 Aug 29]. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON415
- DAI. Haiti declared free of cholera [cited 2023 Aug 29]. https://www.dai.com/news/haiti-declared-free-of-cholera
- Vega Ocasio D, Juin S, Berendes D, Heitzinger K, Prentice-Mott G, Desormeaux AM, et al.; CDC Haiti Cholera Response Group. Cholera Outbreak - Haiti, September 2022-January 2023. MMWR Morb Mortal Wkly Rep. 2023;72:21–5. DOIPubMedGoogle Scholar
- Ministry of Public Health and Population. Epidemiological situation of cholera, 12 May 2023 [in French] [cited 2023 Aug 29]. https://www.mspp.gouv.ht
- Rubin DHF, Zingl FG, Leitner DR, Ternier R, Compere V, Marseille S, et al. Reemergence of Cholera in Haiti. N Engl J Med. 2022;387:2387–9. DOIPubMedGoogle Scholar
- Walters C, Chen J, Stroika S, Katz LS, Turnsek M, Compère V, et al. Genome sequences from a reemergence of Vibrio cholerae in Haiti, 2022 reveal relatedness to previously circulating strains. J Clin Microbiol. 2023;61:
e0014223 . DOIPubMedGoogle Scholar - Ali A, Chen Y, Johnson JA, Redden E, Mayette Y, Rashid MH, et al. Recent clonal origin of cholera in Haiti. Emerg Infect Dis. 2011;17:699–701. DOIPubMedGoogle Scholar
- Azarian T, Ali A, Johnson JA, Mohr D, Prosperi M, Veras NM, et al. Phylodynamic analysis of clinical and environmental Vibrio cholerae isolates from Haiti reveals diversification driven by positive selection. MBio. 2014;5:e01824–14. DOIPubMedGoogle Scholar
- Alam MT, Ray SS, Chun CN, Chowdhury ZG, Rashid MH, Madsen Beau De Rochars VE, et al. Major shift of toxigenic V. cholerae O1 from Ogawa to Inaba serotype isolated from clinical and environmental samples in Haiti. PLoS Negl Trop Dis. 2016;10:
e0005045 . DOIPubMedGoogle Scholar - Paisie TK, Cash MN, Tagliamonte MS, Ali A, Morris JG Jr, Salemi M, et al. Molecular basis of the toxigenic Vibrio cholerae O1 serotype switch from Ogawa to Inaba in Haiti. Microbiol Spectr. 2023;11:
e0362422 . DOIPubMedGoogle Scholar - Weill FX, Domman D, Njamkepo E, Tarr C, Rauzier J, Fawal N, et al. Genomic history of the seventh pandemic of cholera in Africa. Science. 2017;358:785–9. DOIPubMedGoogle Scholar
- Domman D, Quilici ML, Dorman MJ, Njamkepo E, Mutreja A, Mather AE, et al. Integrated view of Vibrio cholerae in the Americas. Science. 2017;358:789–93. DOIPubMedGoogle Scholar
- Colwell RR, Huq A. Environmental reservoir of Vibrio cholerae. The causative agent of cholera. Ann N Y Acad Sci. 1994;740(1 Disease in Ev):44–54.
- Morris JG Jr. Cholera—modern pandemic disease of ancient lineage. Emerg Infect Dis. 2011;17:2099–104. DOIPubMedGoogle Scholar
- Azarian T, Ali A, Johnson JA, Jubair M, Cella E, Ciccozzi M, et al. Non-toxigenic environmental Vibrio cholerae O1 strain from Haiti provides evidence of pre-pandemic cholera in Hispaniola. Sci Rep. 2016;6:36115. DOIPubMedGoogle Scholar
- Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. DOIPubMedGoogle Scholar
- Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:
e15 . DOIPubMedGoogle Scholar - Tonkin-Hill G, Lees JA, Bentley SD, Frost SDW, Corander J. Fast hierarchical Bayesian analysis of population structure. Nucleic Acids Res. 2019;47:5539–49. DOIPubMedGoogle Scholar
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. DOIPubMedGoogle Scholar
- Alam MT, Mavian C, Paisie TK, Tagliamonte MS, Cash MN, Angermeyer A, et al. Emergence and evolutionary response of Vibrio cholerae to novel bacteriophage, Democratic Republic of the Congo. Emerg Infect Dis. 2022;28:2482–90. DOIPubMedGoogle Scholar
- Weill F-X, Domman D, Njamkepo E, Almesbahi AA, Naji M, Nasher SS, et al. Genomic insights into the 2016-2017 cholera epidemic in Yemen. Nature. 2019;565:230–3. DOIPubMedGoogle Scholar
- Sagulenko P, Puller V, Neher RA. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:
vex042 . DOIPubMedGoogle Scholar - Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. DOIPubMedGoogle Scholar
- Yu GC, Smith DK, Zhu HC, Guan Y, Lam TTY. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. DOIGoogle Scholar
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. DOIPubMedGoogle Scholar
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLOS Comput Biol. 2009;5:
e1000520 . DOIPubMedGoogle Scholar - Xie W, Lewis PO, Fan Y, Kuo L, Chen MH. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst Biol. 2011;60:150–60. DOIPubMedGoogle Scholar
- Sjölund-Karlsson M, Reimer A, Folster JP, Walker M, Dahourou GA, Batra DG, et al. Drug-resistance mechanisms in Vibrio cholerae O1 outbreak strain, Haiti, 2010. Emerg Infect Dis. 2011;17:2151–4. DOIPubMedGoogle Scholar
- Kim HB, Wang M, Ahmed S, Park CH, LaRocque RC, Faruque AS, et al. Transferable quinolone resistance in Vibrio cholerae. Antimicrob Agents Chemother. 2010;54:799–803. DOIPubMedGoogle Scholar
- Jubair M, Morris JG Jr, Ali A. Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype. PLoS One. 2012;7:
e45187 . DOIPubMedGoogle Scholar - Haiti Libre. Haiti—FLASH: first effect of Hurricane Fiona on Haiti [cited 2023 Aug 29]. https://www.haitilibre.com/en/news-37687-haiti-flash-first-effect-of-hurricane-fiona-on-haiti.html
- United Nations. ‘Violent civil unrest’ in Haiti hampers aid delivery, 16 Sep 2022 [cited 2023 Aug 29]. https://news.un.org/en/story/2022/09/1126861
- The Guardian. They have no fear and no mercy: gang rule engulfs Haitian capital, 18 Sep 2022 [cited 2023 Aug 29]. https://www.theguardian.com/world/2022/sep/18/haiti-violence-gang-rule-port-au-prince
- Ali M, Emch M, Park JK, Yunus M, Clemens J. Natural cholera infection-derived immunity in an endemic setting. J Infect Dis. 2011;204:912–8. DOIPubMedGoogle Scholar
- Kirpich A, Weppelmann TA, Yang Y, Ali A, Morris JG Jr, Longini IM. Cholera transmission in Ouest Department of Haiti: dynamic modeling and the future of the epidemic. PLoS Negl Trop Dis. 2015;9:
e0004153 . DOIPubMedGoogle Scholar - Kirpich A, Weppelmann TA, Yang Y, Morris JG Jr, Longini IM Jr. Controlling cholera in the Ouest Department of Haiti using oral vaccines. PLoS Negl Trop Dis. 2017;11:
e0005482 . DOIPubMedGoogle Scholar - Sévère K, Rouzier V, Anglade SB, Bertil C, Joseph P, Deroncelay A, et al. Effectiveness of oral cholera vaccine in Haiti: 37-month follow-up. Am J Trop Med Hyg. 2016;94:1136–42. DOIPubMedGoogle Scholar
- Pape JW, Rouzier V. Embracing oral cholera vaccine—shifting response to cholera. N Engl J Med. 2014;370:2067–9. DOIPubMedGoogle Scholar
- Ratnayake R, Finger F, Azman AS, Lantagne D, Funk S, Edmunds WJ, et al. Highly targeted spatiotemporal interventions against cholera epidemics, 2000-19: a scoping review. Lancet Infect Dis. 2021;21:e37–48. DOIPubMedGoogle Scholar
1These senior authors contributed equally to this article.