Volume 29, Number 3—March 2023
Research Letter
New Postmortem Perspective on Emerging SARS-CoV-2 Variants of Concern, Germany
Figure
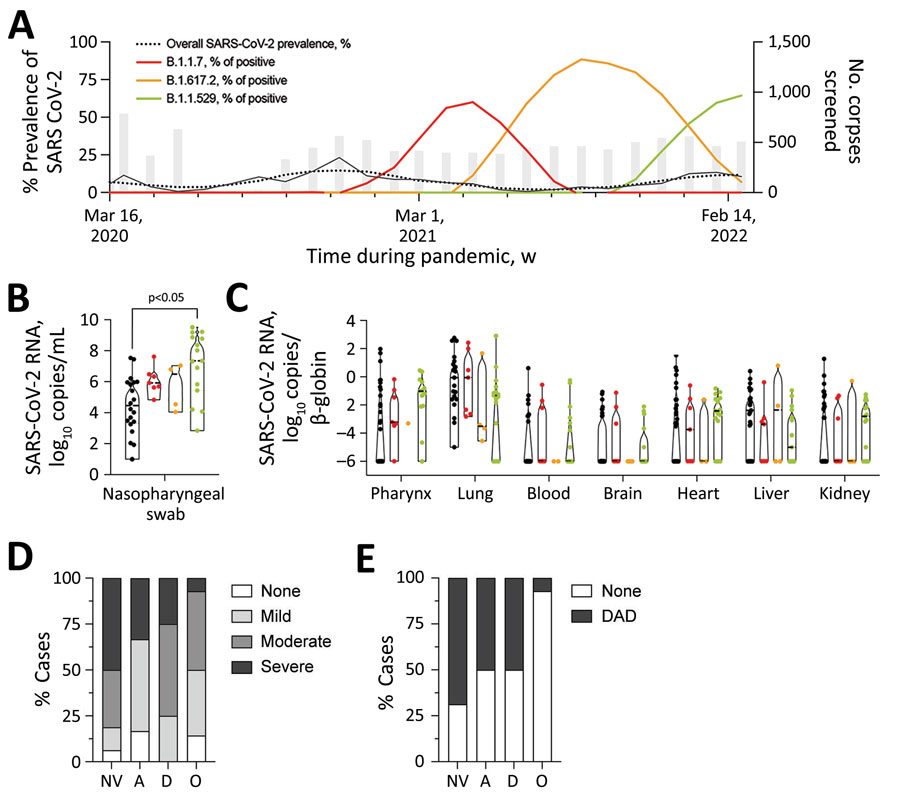
Figure. Prevalence of SARS-CoV-2 variants, pulmonary inflammation, and diffuse alveolar damage in corpses autopsied during 2020 and 2022 at the Institute of Legal Medicine and crematoria in Hamburg, Germany. A) Overall prevalence of corpses positive for SARS-CoV-2 mRNA and prevalence of B.1.1.7 (Alpha), B.1.617.2 (Delta), and B.1.1.529 (Omicron) variants of concern as the percentage of SARS-CoV-2 mRNA-positive corpses are depicted; 3-point centered moving averages are shown. Gray bars indicate monthly number of corpses screened for SARS-CoV-2 mRNA. B, C) Number of SARS-CoV-2 mRNA copies in different autopsy specimens. Median and interquartile ranges of viral mRNA loads were stratified according to virus variants in nasopharyngeal swabs (B) and different organs (C). Nasopharyngeal and organ viral loads for non-VOC and B.1.1.7 were published in part previously (7,8). Black dots are non-VOC lineages. Pairwise comparisons were performed by using the Kruskal-Wallis H test and Dunn post-hoc analysis. D, E) Percentage of cases that had pulmonary inflammation or alveolar damage caused by different SARS-CoV-2 lineages: NV, Non-VOC lineage; A, Alpha B.1.1.7 lineage; D, Delta B.1.617.2 lineage; O, Omicron B.1.1.529 lineage. D) We scored microscopic pulmonary inflammation as follows: none, 0; mild, 1; moderate, 2; or severe, 3 on the basis of a Likert-like scale and calculated percentages of cases within each group for each SARS-CoV-2 variant. E) Percentage of corpses infected with non-VOC lineages (n = 16) and the VOC lineages B.1.1.7 (Alpha, n = 6), B.1.617.2 (Delta, n = 4), and B.1.1.529 (Omicron, n = 14) that had DAD in lungs. DAD, diffuse alveolar damage; VOC, variant of concern.
References
- Choi JY, Smith DM. SARS-CoV-2 variants of concern. Yonsei Med J. 2021;62:961–8. DOIPubMedGoogle Scholar
- Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. 2022;376:
eabn4947 . DOIPubMedGoogle Scholar - Heinrich F, Romich C, Zimmermann T, Kniep I, Fitzek A, Steurer S, et al. Dying of VOC-202012/01 - multimodal investigations in a death case of the SARS-CoV-2 variant. Int J Legal Med. 2022;136:193–202. DOIPubMedGoogle Scholar
- Nörz D, Grunwald M, Olearo F, Fischer N, Aepfelbacher M, Pfefferle S, et al. Evaluation of a fully automated high-throughput SARS-CoV-2 multiplex qPCR assay with built-in screening functionality for del-HV69/70- and N501Y variants such as B.1.1.7. J Clin Virol. 2021;141:
104894 . DOIPubMedGoogle Scholar - Nörz D, Grunwald M, Tang HT, Weinschenk C, Günther T, Robitaille A, et al. Clinical evaluation of a fully-automated high-throughput multiplex screening-assay to detect and differentiate the SARS-CoV-2 B.1.1.529 (Omicron) and B.1.617.2 (Delta) lineage variants. Viruses. 2022;14:608. DOIPubMedGoogle Scholar
- Krasemann S, Dittmayer C, von Stillfried S, Meinhardt J, Heinrich F, Hartmann K, et al. Assessing and improving the validity of COVID-19 autopsy studies - A multicentre approach to establish essential standards for immunohistochemical and ultrastructural analyses. EBioMedicine. 2022;83:
104193 . DOIPubMedGoogle Scholar - Puelles VG, Lütgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–2. DOIPubMedGoogle Scholar
- Ondruschka B, Heinrich F, Lindenmeyer M, Edler C, Möbius D, Czogalla J, et al. Multiorgan tropism of SARS-CoV-2 lineage B.1.1.7. Int J Legal Med. 2021;135:2347–9. DOIPubMedGoogle Scholar
- Walsh KA, Jordan K, Clyne B, Rohde D, Drummond L, Byrne P, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81:357–71. DOIPubMedGoogle Scholar
- Mohandas S, Yadav PD, Sapkal G, Shete AM, Deshpande G, Nyayanit DA, et al. Pathogenicity of SARS-CoV-2 Omicron (R346K) variant in Syrian hamsters and its cross-neutralization with different variants of concern. EBioMedicine. 2022;79:
103997 . DOIPubMedGoogle Scholar
1These authors contributed equally to this article.