Volume 29, Number 3—March 2023
Research Letter
New Postmortem Perspective on Emerging SARS-CoV-2 Variants of Concern, Germany
Cite This Article
Citation for Media
Abstract
We performed autopsies on persons in Germany who died from COVID-19 and observed higher nasopharyngeal SARS-CoV-2 viral loads for variants of concern (VOC) compared with non-VOC lineages. Pulmonary inflammation and damage appeared higher in non-VOC than VOC lineages until adjusted for vaccination status, suggesting COVID-19 vaccination may mitigate pulmonary damage.
SARS-CoV-2 emerged in 2020, and after rapid global spread, virus variants emerged that had adaptation or immune evasion mutations and were classified as variants of concern (VOCs). Although the first VOCs, Alpha (B.1.1.7) and Delta (B.1.617.2), showed enhanced transmissibility (1), Omicron (B.1.1.529) lineages carry mutations that provide strong immune evasion after infection with previous lineages or mRNA vaccination (2). Because autopsy data are lacking, differences in SARS-CoV-2–related pulmonary disease and tropisms have not been well studied. In this study, we performed full autopsies of persons who died from COVID-19 and conducted comprehensive analyses to characterize COVID-19–related cases macroscopically and microscopically.
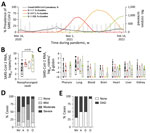
All corpses admitted to the Institute of Legal Medicine (n = 8,578) and crematoria (n = 1,705) in Hamburg, Germany, during March 3, 2020–March 31, 2022, were screened for SARS-CoV-2 mRNA by quantitative reverse transcription PCR (3). We found a total of 808 SARS-CoV-2 RNA-positive corpses; median monthly prevalence was 6.54% in 2020, 5.28% in 2021, and 12.50% in 2022 (Figure, panel A). In comparison, the median monthly prevalence of SARS-CoV-2 in Hamburg’s general population was 0.10% in 2020, 0.37% in 2021, and ≈5.20% in 2022 (https://de.statista.com/statistik/daten/studie/1106006/umfrage/entwicklung-der-fallzahl-des-coronavirus-in-hamburg). A considerably higher prevalence of virus in deceased persons than in the general population can be explained by an older age in postmortem cohorts, targeted transport of clinically suspected or confirmed COVID-19 deaths to the Institute of Legal Medicine in 2020, and underreporting of overall SARS-CoV-2 prevalence because of limited availability of molecular testing.
We further characterized SARS-CoV-2 RNA-positive samples by using multiplexed typing quantitative reverse transcription PCR (4,5). The occurrence of VOCs among corpses (B.1.1.7 for December 2020–March 2022, B.1.617.2 beginning in May 2021, and B.1.1.529 beginning in November 2021) reflected their occurrence within the general population but had a 2–4 week delay (https://www.leibniz-liv.de/de/aktuelles/covid-19/daten-der-hamburg-surveillance-plattform).
Of the 808 COVID-19–associated deaths (determined by postmortem SARS-CoV-2 RNA detection), we autopsied 49 corpses and collected multiorgan tissue samples for more detailed analyses. We included 23/49 consecutive deceased persons infected with non-VOC lineages and 26/49 consecutive deceased persons infected with VOCs (B.1.1.7, n = 7; B.1.617.2, n = 4; and B.1.1.529, n = 15) (Table). We processed formalin-fixed paraffin-embedded tissue for histologic and immunohistochemical staining and cryopreserved tissue for molecular analysis as previously described (3,6).
The median nasopharyngeal SARS-CoV-2 RNA load was 5.82 (interquartile range [IQR] 4.08–7.31) log10 copies/mL (Figure, panel B). Nasopharyngeal and organ viral loads for non-VOC and B.1.1.7 were published in part previously (7,8). We observed strong evidence for a difference in mean nasopharyngeal viral loads by virus variant (p = 0.01); by using multiple comparisons, we observed a difference in means between B.1.1.529 and non-VOC lineages (p = 0.002; Figure, panel B). This result supports increased infectivity of B.1.1.529 compared with non-VOC lineages (9). An association between nasopharyngeal virus load and virus variant at the 5% significance level did not persist in a multivariable model adjusted for the deceased’s vaccination status, which might be because of the small sample size (Appendix Tables 1, 2). Of note, the pulmonary virus load was strongly associated with viremia (odds ratio 2.21, 95% CI 1.34–3.63; p = 0.002) and mRNA detection in peripheral organs (odds ratio 1.54, 95% CI 1.10–2.16; p = 0.01) in univariable logistic regression models. However, normalized SARS-CoV-2 RNA loads in peripheral organs did not differ between virus variants (Figure, panel C).
Experienced pathologists performed single-blind histologic assessments. We detected SARS-CoV-2 nucleocapsid protein in the lungs of 25/41 (61%) cases. Using a Likert-like scale, we found the median abundance of SARS-CoV-2 nucleocapsid protein (0, not detected; 1, low abundance; 2, intermediate abundance; 3, high abundance) was 1 (IQR 0–2) for non-VOC lineage, 1.5 (IQR 1–2) for B.1.1.7, 0.5 (IQR 0–1) for B.1.617.2, and 0 (IQR 0–1) for B.1.1.529 cases (p = 0.03) (Appendix Figure).
We detected mild to strong inflammatory changes in the lungs of 36/40 (90%) cases and microscopic signs of diffuse alveolar damage (DAD), indicating acute respiratory distress syndrome, in 17/40 (43%) cases (Figure, panels D, E). As in recent animal experiments (10), pulmonary changes, such as inflammation and DAD, were associated with virus variant at the 5% level (Appendix Tables 3, 4) but not with nasopharyngeal or pulmonary viral load (inflammatory changes, p>0.05; DAD, p>0.05). An association between virus variants and inflammatory changes or DAD at the 5% level did not persist in a multivariable model adjusted for the deceased’s vaccination status (Appendix Tables 5–7).
In conclusion, our data confirm higher SARS-CoV-2 mRNA loads in nasopharynges of deceased persons who were infected with the B.1.1.529 VOC lineage, but we observed no differences in pulmonary or tertiary organ viral loads. However, pulmonary inflammation appeared higher and DAD more frequent in non-VOC than VOC lineages until adjustment for vaccination status. Our results suggest that prior vaccination, rather than reduced virulence of virus variants, might convey protection against pulmonary inflammation and acute respiratory distress syndrome during SARS-CoV-2 infections.
Dr. Heinrich is a medical doctor at the Institute of Legal Medicine, University Medical Center Hamburg-Eppendorf, Germany. His primary research interests focus on infectious disease and immunology.
Acknowledgments
We thank Christiane Stark and Kristin Hartmann for technical support and Daniela Fröb for managing the logistics of SARS-CoV-2–associated incoming corpses within the Institute of Legal Medicine.
We offer condolences to the families and friends of all the patients whose deaths were attributed to COVID-19.
This work was funded by the research consortium NATON. The NATON project (grant no. 01KX2121) is part of the National Network University Medicine funded by the Federal Ministry of Education and Research, Germany. The National Network University Medicine is coordinated at the Charité–Universitätsmedizin Berlin and supervised by the German Aerospace Center (DLR Project Management Agency). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
References
- Choi JY, Smith DM. SARS-CoV-2 variants of concern. Yonsei Med J. 2021;62:961–8. DOIPubMedGoogle Scholar
- Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. 2022;376:
eabn4947 . DOIPubMedGoogle Scholar - Heinrich F, Romich C, Zimmermann T, Kniep I, Fitzek A, Steurer S, et al. Dying of VOC-202012/01 - multimodal investigations in a death case of the SARS-CoV-2 variant. Int J Legal Med. 2022;136:193–202. DOIPubMedGoogle Scholar
- Nörz D, Grunwald M, Olearo F, Fischer N, Aepfelbacher M, Pfefferle S, et al. Evaluation of a fully automated high-throughput SARS-CoV-2 multiplex qPCR assay with built-in screening functionality for del-HV69/70- and N501Y variants such as B.1.1.7. J Clin Virol. 2021;141:
104894 . DOIPubMedGoogle Scholar - Nörz D, Grunwald M, Tang HT, Weinschenk C, Günther T, Robitaille A, et al. Clinical evaluation of a fully-automated high-throughput multiplex screening-assay to detect and differentiate the SARS-CoV-2 B.1.1.529 (Omicron) and B.1.617.2 (Delta) lineage variants. Viruses. 2022;14:608. DOIPubMedGoogle Scholar
- Krasemann S, Dittmayer C, von Stillfried S, Meinhardt J, Heinrich F, Hartmann K, et al. Assessing and improving the validity of COVID-19 autopsy studies - A multicentre approach to establish essential standards for immunohistochemical and ultrastructural analyses. EBioMedicine. 2022;83:
104193 . DOIPubMedGoogle Scholar - Puelles VG, Lütgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–2. DOIPubMedGoogle Scholar
- Ondruschka B, Heinrich F, Lindenmeyer M, Edler C, Möbius D, Czogalla J, et al. Multiorgan tropism of SARS-CoV-2 lineage B.1.1.7. Int J Legal Med. 2021;135:2347–9. DOIPubMedGoogle Scholar
- Walsh KA, Jordan K, Clyne B, Rohde D, Drummond L, Byrne P, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81:357–71. DOIPubMedGoogle Scholar
- Mohandas S, Yadav PD, Sapkal G, Shete AM, Deshpande G, Nyayanit DA, et al. Pathogenicity of SARS-CoV-2 Omicron (R346K) variant in Syrian hamsters and its cross-neutralization with different variants of concern. EBioMedicine. 2022;79:
103997 . DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: February 14, 2023
1These authors contributed equally to this article.
Table of Contents – Volume 29, Number 3—March 2023
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Fabian Heinrich, Center for Diagnostics, Institute of Legal Medicine, University Medical Center Hamburg Eppendorf, Butenfeld 34, 20259 Hamburg, Germany
Top