SARS-CoV-2 Spillback to Wild Coatis in Sylvatic–Urban Hotspot, Brazil
Ana Gabriella Stoffella-Dutra, Bruna Hermine de Campos, Pedro Henrique Bastos e Silva, Karolina Lopes Dias, Iago José da Silva Domingos, Nadja Simbera Hemetrio, Joilson Xavier, Felipe Iani, Vagner Fonseca, Marta Giovanetti, Leonardo Camilo de Oliveira, Mauro Martins Teixeira, Zelia Ines Portela Lobato, Helena Lage Ferreira, Clarice Weis Arns, Edison Durigon, Betânia Paiva Drumond, Luiz Carlos Junior Alcantara, Marcelo Pires Nogueira de Carvalho, and Giliane de Souza Trindade

Author affiliations: Universidade Federal de Minas Gerais, Belo Horizonte, Brazil (A.G. Stoffella-Dutra, B.H. de Campos, P.H.B. e Silva, K.L. Dias, I.J.S. Domingos, J. Xavier, L.C. de Oliveira, M.M. Teixeira, Z.I.P. Lobato, B.P. Drumond, M.P.N. de Carvalho, G. de Souza Trindade); Fundação de Parques Municipais e Zoobotânica de Belo Horizonte, Belo Horizonte (N.S. Hemetrio); Fundação Ezequiel Dias, Belo Horizonte (F. Iani); Organização Pan-Americana da Saúde, Brasília, Brazil (V. Fonseca); University of Campus Bio-Medico di Roma, Rome, Italy (M. Giovanetti); Fundação Oswaldo Cruz, Rio de Janeiro, Brazil (M. Giovanetti, L.C.J. Alcantara); Universidade de São Paulo, São Paulo, Brazil (H.L. Ferreira, E. Durigon); Universidade de Campinas, Campinas, Brazil (C.W. Arns)
Main Article
Figure
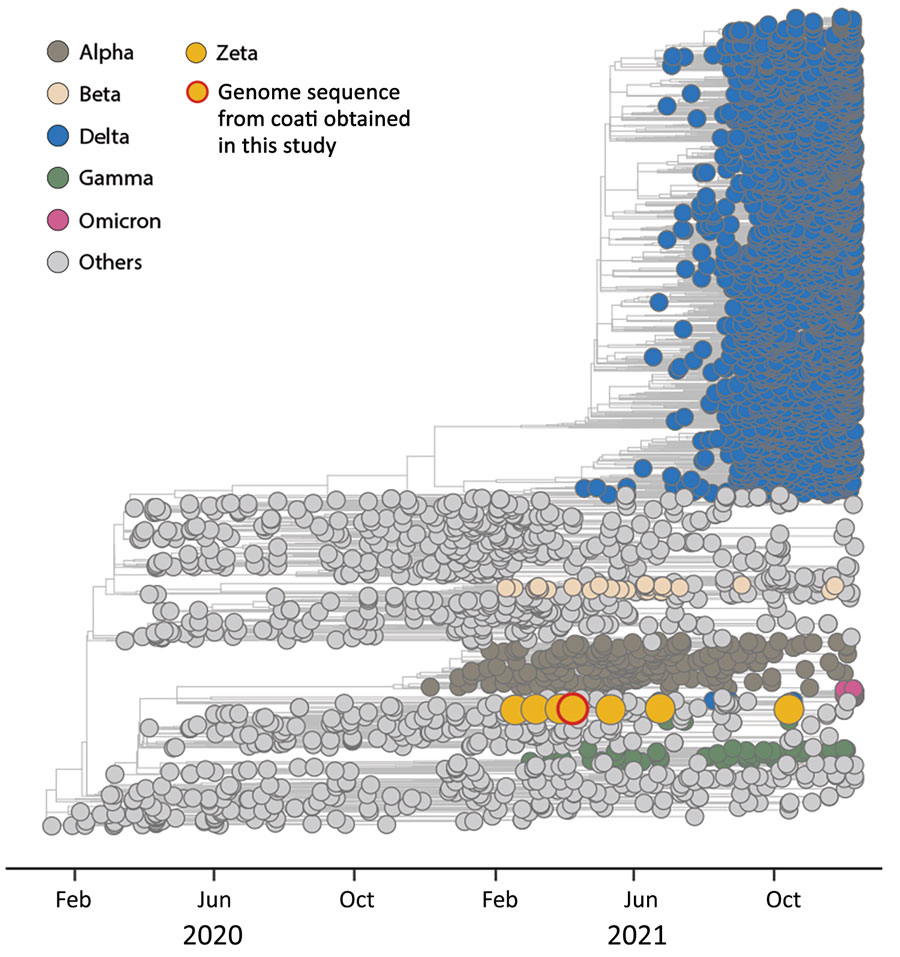
Figure. Time-scaled phylogenetic analysis of SARS-CoV-2 sequences, by variant type, in study of SARS-CoV-2 spillback to wild coatis in sylvatic–urban hotspot, Brazil. Maximum-likelihood method was used to compare the complete genomic sequence of SARS-CoV-2 obtained from coati (Nasua nasua) 535 (red-outlined yellow circle) and 3,441 SARS-CoV-2 reference genomic sequences (GISAID, https://www.gisaid.org) from around the world collected through October 2021. Colors represent clades corresponding to different SARS-CoV-2 variants of concern described by the World Health Organization; yellow indicates Zeta variant sequences. The SARS-CoV-2 sequence generated in this study was deposited in the GISAID database (accession no. EPI_ISL_8800460) and SisGen (Sistema Nacional de Gestão do Patrimônio Genético, https://www.sisgen.gov.br; no. A627307).
Main Article
Page created: January 12, 2023
Page updated: February 20, 2023
Page reviewed: February 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
