Spatiotemporal Evolution of SARS-CoV-2 Alpha and Delta Variants during Large Nationwide Outbreak of COVID-19, Vietnam, 2021
Nguyen Thi Tam
1, Nguyen To Anh
1
, Trinh Son Tung
1, Pham Ngoc Thach, Nguyen Thanh Dung, Van Dinh Trang, Le Manh Hung, Trinh Cong Dien, Nghiem My Ngoc, Le Van Duyet, Phan Manh Cuong, Hoang Vu Mai Phuong, Pham Quang Thai, Nguyen Le Nhu Tung, Dinh Nguyen Huy Man, Nguyen Thanh Phong, Vo Minh Quang, Pham Thi Ngoc Thoa, Nguyen Thanh Truong, Tran Nguyen Phuong Thao, Dao Phuong Linh, Ngo Tan Tai, Ho The Bao, Vo Trong Vuong, Huynh Thi Kim Nhung, Phan Nu Dieu Hong, Le Thi Phuoc Hanh, Le Thanh Chung, Nguyen Thi Thanh Nhan, Ton That Thanh, Do Thai Hung, Huynh Kim Mai, Trinh Hoang Long, Nguyen Thu Trang, Nguyen Thi Hong Thuong, Nguyen Thi Thu Hong, Le Nguyen Truc Nhu, Nguyen Thi Han Ny, Cao Thu Thuy, Le Kim Thanh, Lam Anh Nguyet, Le Thi Quynh Mai, Tang Chi Thuong, Le Hong Nga, Tran Tan Thanh, Guy Thwaites, H. Rogier van Doorn, Nguyen Van Vinh Chau, Thomas Kesteman, Le Van Tan

, and
for the OUCRU COVID-19 Research Group2
Author affiliations: Oxford University Clinical Research Unit, Hanoi, Vietnam (N.T. Tam, T.S. Tung, N.T. Trang, N.T.H. Thuong, T. Kesteman, H. R. van Doorn); National Hospital for Tropical Diseases, Hanoi (P.N. Thach, V.D. Trang, L.V. Duyet, P.M. Cuong); Vietnam Military Medical University, Hanoi (T.C. Dien); National Institute of Hygiene and Epidemiology, Hanoi (H.V.M. Phuong, L.T.Q. Mai, P.Q. Thai); Hue National Hospital, Hue, Vietnam (P.N.D. Hong, L.T.P. Hanh); Da Nang Center for Disease Control, Da Nang, Vietnam (L.T. Chung, N.T.T. Nhan, T.T. Thanh); Pasteur Institute, Nha Trang, Khanh Hoa, Vietnam (D.T. Hung, H.K. Mai, T.H. Long); Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam (N.T. Anh, N.T.T. Hong, L.N.T. Nhu, N.T.H. Ny, T.T. Thanh, C.T. Thuy, L.K. Thanh, L.A. Nguyet, G. Thwaites, L.V. Tan); Hospital for Tropical Diseases, Ho Chi Minh City (N.T. Dung, L.M. Hung, N.L.N. Tung, D.N.H. Man, N.M. Ngoc, N.T. Phong, V.M. Quang, P.T.N. Thoa, N.T. Truong, T.N.P. Thao, D.P. Linh, N.T. Tam, H.T. Bao, V.T. Vuong, H.T.K. Nhung); Department of Health, Ho Chi Minh City, (N.V.V. Chau, T.C. Thuong); Ho Chi Minh City Center for Disease Control, Ho Chi Minh City (L.H. Nga); University of Oxford, Oxford, UK (H.R. van Doorn, G. Thwaites, L.V. Tan)
Main Article
Figure 1
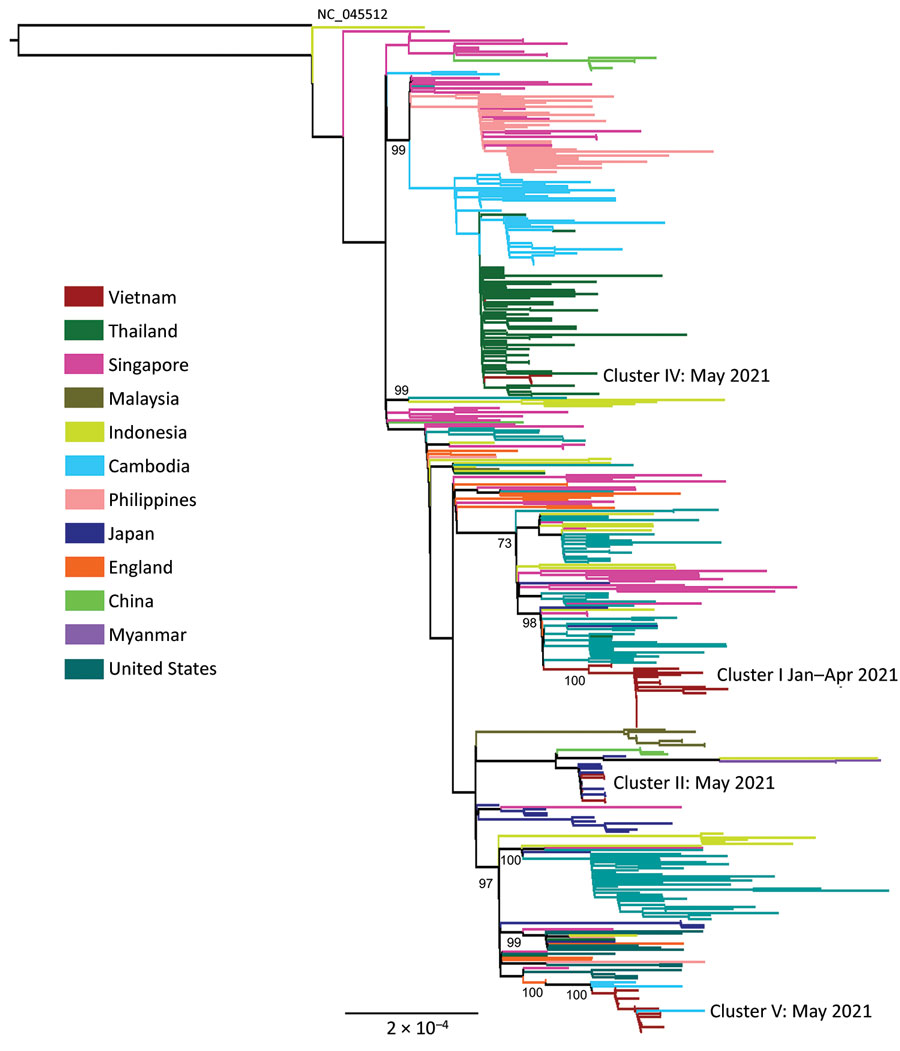
Figure 1. Maximum-likelihood tree of SARS-CoV-2 Alpha and Delta variants during large nationwide outbreak of COVID-19, Vietnam, 2021. Complete coding sequences of Alpha variant viruses circulating in Vietnam in 2021 are shown. Phylogenetic clusters were named accordingly to community clusters recorded during the study period shown in Appendix Figure 1). Numbers along branches are bootstrap values. Cluster II was linked with a traveler from Japan (Appendix Figure 1). Outgroup was the SARS-CoV-2 wild-type strain (GenBank accession no. NC_0455122).
Main Article
Page created: March 08, 2023
Page updated: April 19, 2023
Page reviewed: April 19, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
