Volume 29, Number 5—May 2023
Dispatch
Rustrela Virus as Putative Cause of Nonsuppurative Meningoencephalitis in Lions
Figure 2
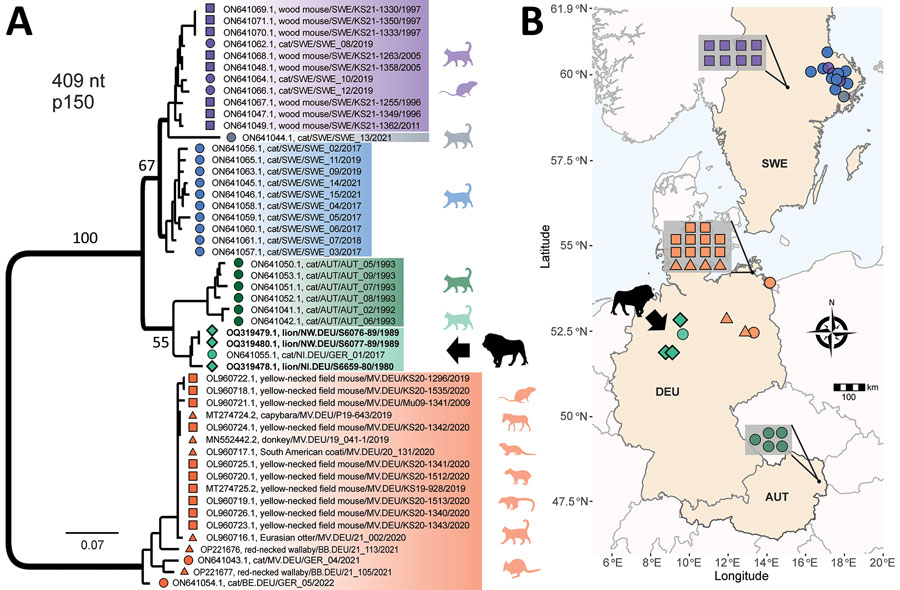
Figure 2. Phylogenetic analysis and spatial distribution of rustrela virus infections in Europe. A) Maximum-likelihood phylogenetic tree of partial rustrela virus (RusV) sequences (409 nt, representing genome positions 100–508 of donkey-derived RusV reference genome, GenBank accession no. MN552442.2). Only bootstrap values at major branches are shown in the phylogenetic tree. RusV sequence names are shown in the format host/ISO 1366 code of location (federal, state, country)/animal ID/year. The tree was produced using IQ-TREE version 2.2.0; transition model 2 plus empirical frequency plus gamma 4 with 1,000 bootstrap replicates. Bold text indicates sequences from this study. Scale bar indicates substitutions per site. B) Mapping of the geographic origin of RusV-positive animals in Europe. Colors represent the phylogenetic clades of the sequences. Diamonds represent lions; circles, domestic cats; triangles, other zoo animals; squares, Apodemus spp. rodents. Symbols in gray boxes represent individuals from the same or very close locations. AUT, Austria; BE, Berlin; DEU/GER, Germany; MV, Mecklenburg–Western Pomerania; NI, Lower Saxony; NW, North Rhine–Westphalia; SWE, Sweden.