Volume 29, Number 6—June 2023
Dispatch
Detection of Novel Poxvirus from Gray Seal (Halichoerus grypus), Germany
Cite This Article
Citation for Media
Abstract
We detected a novel poxvirus from a gray seal (Halichoerus grypus) from the North Sea, Germany. The juvenile animal showed pox-like lesions and deteriorating overall health condition and was finally euthanized. Histology, electron microscopy, sequencing, and PCR confirmed a previously undescribed poxvirus of the Chordopoxvirinae subfamily, tentatively named Wadden Sea poxvirus.
Members of the poxvirus subfamily Chordopoxvirinae (family Poxviridae) infect vertebrates, such as birds, reptiles, and a broad spectrum of mammals. Although some chordopoxviruses have a narrow host range, several can easily jump species barriers and cause severe disease (1). Considering the potential zoonotic and epizootic potential of chordopoxviruses, constant monitoring and adaptation of diagnostic procedures are essential. With the advent of metagenomic sequencing, novel chordopoxviruses have been identified that are genetically diverse and were not readily detectable by using established PCR-based diagnostics (2–4).
We report a case of a poxvirus infection in a gray seal (Halichoerus grypus) from the North Sea near Germany. We identified a novel chordopoxvirus that was phylogenetically divergent from other known poxviruses of gray seals.
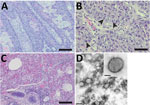
In June 2020, a juvenile gray seal was nursed at a rehabilitation center in Friedrichskoog, Germany (Appendix Figure 1) and was about to be released back into the wild when staff noticed pox-like lesions on its hind flipper (Appendix Figure 2). The seal’s overall health condition deteriorated over the next 3 weeks, and it had dyspnea, vomiting after feeding, and apathy; it was humanely euthanized. At necropsy, the seal was in good body condition (abdominal blubber ≈35 mm, reference >30 mm). We noted 2 prominent verrucous nodules on the right hind flipper (Appendix Figure 2). We also found severe emphysema of the mediastinum and a focal, adhesive pleuritis. No other organs had lesions. Histologic examination of both nodules (Appendix) revealed severe papillary epithelial hyperplasia, acanthosis, ballooning degeneration, large eosinophilic cytoplasmatic inclusion bodies in the stratum spinosum, moderate hyperkeratosis, and severe ulceration with hemorrhages (Figure 1, panels A, B). In addition, we observed multifocal severe infiltrations of neutrophils. In the liver, we detected a focally necrotizing hepatitis with ballooning degeneration of nuclei and a focal granulomatous subcapsular hepatitis with intralesional parasites and calcification. We found further inflammatory changes in the lungs (Figure 1, panel C), which had multifocal moderate pneumonia with infiltration of mononuclear cells and neutrophils; the heart had focal severe mononuclear myocarditis; and the duodenum had moderate diffuse lymphoplasmacellular enteritis. We also observed depletion of lymphocytic organs, including a severe atrophy of the thymus.
Results of quantitative PCRs (qPCRs) specific for orthopoxvirus (5) and parapoxvirus (6), canine alphaherpesvirus 1, influenza A virus, canine morbillivirus, and Brucella sp. were negative for lung and skin lesion tissue. We isolated an Escherichia coli strain from lung, mediastinum, liver, kidney, and intestines.
We used electron microscopy to analyze lung tissue and detected typical poxvirus-like virions, which were ovoid in shape and ≈250 nm long and ≈200 nm wide (Figure 1, panel D). The surface structures resembled typical orthopox-like randomly arranged tubular units. However, the virion morphology did not enable assignment to a poxvirus genus.
We isolated DNA from a pool of lung and skin lesion tissue and sequenced DNA using Ion Torrent S5XL (Thermo Fisher Scientific, https://www.thermofisher.com) (7), NovaSeq (Illumina, https://www.illumina.com), and MinION Mk1C (Oxford Nanopore Technologies, https://nanoporetech.com) sequencing technologies (Appendix). We combined the reads in a hybrid assembly, which resulted in a complete poxviral genome (mean coverage ≈650). We were able to confirm completeness of the genome because the terminal repeats contained the terminal hairpin region.
We screened several organs by using 2 different virus-specific qPCRs (Appendix). Results from both qPCR panels were consistent and we detected the highest viral loads in the skin lesion and parts of the lung (Table).
We tentatively named the poxvirus Wadden Sea poxvirus (WSPV) to reflect the geographic origin of the infected gray seal, which was found in the Wadden Sea, an intertidal zone in the southeastern part of the North Sea, Germany (Appendix Figure 1). We submitted the annotated WSPV genome sequence to the International Nucleotide Sequence Database Collaboration (https://www.insdc.org; accession no. OP810554).
WSPV had one of the smallest genomes (124,614 bp) and lowest guanine-cytosine content (≈22.5%) described so far among chordopoxviruses. The unique core genome of 117,842 bp was flanked by 2 inverted terminal repeats of 3,386 bp each. We identified 124 unique potential open reading frames (ORFs), of which 3 were duplicated in the inverted terminal repeats. BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi) identified 111 ORFs representing orthologs of poxvirus proteins. Nine ORFs encoded proteins that were not related to known poxvirus proteins but showed sequence similarity to eukaryote proteins, and 4 ORFs remained unclassified.
For phylogenetic classification, we compared the amino acid sequences encoded by 15 poxvirus core genes from WSPV with the respective homologs from 47 representative poxviruses (Appendix). WSPV formed a separate phylogenetic branch that did not fall within any of the established genera (Figure 2) and likely is a new species within a novel genus of the subfamily Chordopoxvirinae. Of note, a sequence comparison of the predicted WSPV DNA polymerase protein to the nonredundant BLAST database revealed a 96.3% sequence identity with a partial sequence from a Steller sea lion poxvirus (GenBank accession no. AAR06586.1), but other poxviruses had a sequence identity <77%.
Infections with poxviruses have been reported from gray seals and harbor seals (Phoca vitulina), both of which live in the North Sea, Germany. Poxviruses have also been reported in other pinniped species and infections are usually associated to subclades of parapoxviruses, called sealpox or sea lion pox virus (8–10). Ulcerative to proliferative, nodular, cutaneous, and mucosal lesions have been found in seals infected with parapoxviruses (10–12). The nodules usually heal spontaneously, but healing lasts from several weeks up to a few months. The illness rate is high, but death rates are low (13). Rarely, nodules in the oral cavity can lead to problems during feeding, and secondary bacterial infection can fatally impair respiratory functions (14). However, in the case we report, we could not link the animal’s overall deteriorating health condition and severe pneumonia to oral lesions. We considered the cultured E. coli strain a facultative pathogen that might have been involved in disease; however, postmortem contamination might be more likely.
We did not detect any parapoxvirus DNA, but sequencing revealed WSPV, a novel poxvirus that is phylogenetically distinct from any other members of the subfamily Chordopoxvirinae. The high loads of viral DNA in several organs (Table), and the observed histopathologic changes suggested a generalized infection with systemic pathology. Lesion associated detection of high viral loads (cycle quantification [cq] values for skin cq ≈9, for lung cq ≈18) indicated that the WSPV infection was likely responsible for the gray seal’s disease and severe pneumonia. However, other factors might have been involved, and the source of infection, the potential natural reservoir, and the zoonotic potential of WSPV are unknown. None of the contact animals within the rehabilitation center had similar lesions develop and so far, no further cases have been reported.
Sequence comparison of WSPV showed that a close relative of this novel poxvirus has been detected in a cutaneous lesion of a young Steller sea lion (Eumetopias jubatus) from Prince William Sound, Alaska, USA (15). This finding suggests a geographically wide distribution of WSPV or WSPV-related viruses and the potential to infect other pinnipeds. As noted in the case we describe, WSPV can cause severe disease. Therefore, future diagnostic considerations for pox-like lesions of pinniped species should include WSPV.
Dr. Pfaff is a biotechnologist and the head of the Laboratory for Applied Bioinformatics and Sequencing of Viral Genomes and Transcriptomes at the Friedrich-Loeffler-Institut, Greifswald, Germany. His main research interests are discovery, characterization, and classification of viruses using high-throughput sequencing and bioinformatics.
Acknowledgment
We gratefully acknowledge Patrick Zitzow for excellent technical assistance with metagenomic sequencing and Mandy Jörn for the graphic design of electron microscope pictures.
References
- Hodo CL, Mauldin MR, Light JE, Wilkins K, Tang S, Nakazawa Y, et al. Novel poxvirus in proliferative lesions of wild rodents in east central Texas, USA. Emerg Infect Dis. 2018;24:1069–72. DOIPubMedGoogle Scholar
- David D, Davidson I, Berkowitz A, Karniely S, Edery N, Bumbarov V, et al. A novel poxvirus isolated from an Egyptian fruit bat in Israel. Vet Med Sci. 2020;6:587–90. DOIPubMedGoogle Scholar
- Wibbelt G, Tausch SH, Dabrowski PW, Kershaw O, Nitsche A, Schrick L. Berlin squirrelpox virus, a new poxvirus in red squirrels, Berlin, Germany. Emerg Infect Dis. 2017;23:1726–9. DOIPubMedGoogle Scholar
- Olson VA, Laue T, Laker MT, Babkin IV, Drosten C, Shchelkunov SN, et al. Real-time PCR system for detection of orthopoxviruses and simultaneous identification of smallpox virus. J Clin Microbiol. 2004;42:1940–6. DOIPubMedGoogle Scholar
- Nitsche A, Büttner M, Wilhelm S, Pauli G, Meyer H. Real-time PCR detection of parapoxvirus DNA. Clin Chem. 2006;52:316–9. DOIPubMedGoogle Scholar
- Wylezich C, Papa A, Beer M, Höper D. A versatile sample processing workflow for metagenomic pathogen detection. Sci Rep. 2018;8:13108. DOIPubMedGoogle Scholar
- Tryland M, Klein J, Nordøy ES, Blix AS. Isolation and partial characterization of a parapoxvirus isolated from a skin lesion of a Weddell seal. Virus Res. 2005;108:83–7. DOIPubMedGoogle Scholar
- Costa H, Klein J, Breines EM, Nollens HH, Matassa K, Garron M, et al. A comparison of parapoxviruses in North American pinnipeds. Front Vet Sci. 2021;8:
653094 . DOIPubMedGoogle Scholar - Günther T, Haas L, Alawi M, Wohlsein P, Marks J, Grundhoff A, et al. Recovery of the first full-length genome sequence of a parapoxvirus directly from a clinical sample. Sci Rep. 2017;7:3734. DOIPubMedGoogle Scholar
- Roess AA, Levine RS, Barth L, Monroe BP, Carroll DS, Damon IK, et al. Sealpox virus in marine mammal rehabilitation facilities, North America, 2007-2009. Emerg Infect Dis. 2011;17:2203–8. DOIPubMedGoogle Scholar
- Müller G, Gröters S, Siebert U, Rosenberger T, Driver J, König M, et al. Parapoxvirus infection in harbor seals (Phoca vitulina) from the German North Sea. Vet Pathol. 2003;40:445–54. DOIPubMedGoogle Scholar
- Hicks BD, Worthy GA. Sealpox in captive grey seals (Halichoerus grypus) and their handlers. J Wildl Dis. 1987;23:1–6. DOIPubMedGoogle Scholar
- Tryland M. Seal parapoxvirus. In: Liu D, editor. Molecular detection of human viral pathogens, 1st edition. New York: CRC Press; 2010. p. 1029–38.
- Bracht AJ, Brudek RL, Ewing RY, Manire CA, Burek KA, Rosa C, et al. Genetic identification of novel poxviruses of cetaceans and pinnipeds. Arch Virol. 2006;151:423–38. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 29, Number 6—June 2023
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Martin Beer, Friedrich-Loeffler-Institut, Bundesforschungsinstitut für Tiergesundheit, Südufer 10, Greifswald 17493, Germany
Top