Volume 29, Number 7—July 2023
Dispatch
Detecting, Quantifying, and Isolating Monkeypox Virus in Suspected Cases, Spain
Cite This Article
Citation for Media
Abstract
When a monkeypox virus outbreak began in several parts of the world in May 2022, timely and accurate diagnosis became mandatory. In our laboratory, a real-time quantitative PCR was designed and evaluated in several patient samples and compared with isolation results. Genomic viral load was related to virus viability.
After smallpox was eradicated worldwide in 1980 and routine smallpox vaccination subsequently ceased, monkeypox virus (MPXV) emerged as the most consequential orthopoxvirus for public health (1). The genetic clades of MPXV are clade I (formerly the Congo Basin [Central African] clade), which is associated with higher virulence and greater mortality rate, and clade II (formerly the West African clade) (1,2).
In May 2022, the United Kingdom reported an outbreak of mpox (formerly monkeypox) that subsequently spread globally (3); Spain was one of the most affected countries (4). The rapid increase in mpox cases has challenged clinical laboratories to understand the spread and transmission of MPXV. We sought to detect and isolate MPXV in persons at risk for infection by genomic amplification using a real-time quantitative PCR (qPCR) and isolation in culture cell.
This study did not require research ethics committee approval because it describes analyses that were completed at the public laboratory as part of routine clinical testing and surveillance during the mpox outbreak in Asturias in the northwest of Spain. Therefore, this study was considered public health practice and was exempt from this requirement.
During May 24–July 15, 2022, a total of 66 samples (42 lesion swabs, 20 respiratory samples, and 4 blood samples) belonging to 41 adults (mean age 35.61 ± 11.25, range 17–60 years) and collected within 72 hours of illness onset were submitted in accordance with laboratory requirements (Table 1). All patients were located in a 30-km area around Oviedo, the capital of Asturias. In addition to MPXV, we also tested for herpes simplex virus, varicella zoster virus, enterovirus, human herpesvirus 8, molluscum contagiosum virus, and human papilloma virus, according to clinical manifestations. Samples were processed following laboratory protocols for nucleic acid detection, and 21 were inoculated onto monolayer conventional cell culture (MRC-5 cell, Vero-E6, and A549 and LLC-MK2 cells subcultures).
We extracted nucleic acids by using the automated nucleic acid purifier Magnapure 96 (Roche Diagnostics, https://www.roche.com). We performed orthopoxvirus group PCR (5) and specific MPXV in-house real-time qPCR.
For qPCR, we amplified 5 µL of extracted nucleic acids in a final volume of 10 µL, including the Brillant III Ultra-fast QPCR Master MIX (Agilent Technologies, https://www.agilent.com), 1,000 nm of each primer (MPXV-S, TGTTGACGCACCAGCGTCT; MPXV-A, AACAGTGGACCCTTGATGACTGT), and 200 nm of FAM-labeled MGB probe (CAATCCATGGTATTCGA; ABI, CA). We performed qPCR as follows: 95° for 7 min, 45 cycles of 95° for 5 min and 60° for 33 min. In addition, we quantified the human β-globin gene in each sample to evaluate sample quality and to calculate normalized viral load in log10 copies per 103 cells (6).
We detected MPXV in 23 (56.09%) of the 41 patients studied. All were men; the mean age was 38.13 + 9.42 years (range 22–56 years). At least 15 stated they had sex with men, and 12 were HIV-positive. We detected herpes simplex virus 1 in 2 persons (1 of whom was co-infected with MPXV) and varicella zoster virus 1 person. MPXV was detected in 25 (96.15%) lesion swab samples with a viral load of 7.58 + 2.03 (range 3.84–11.71 [95% CI 6.44–8.19]) and in 8 (66.66%) respiratory swab specimens with a viral load of 5.04 + 1.01 (range 3.5–6.23 [95% CI 4.192–5.88]; p = 0.0041).
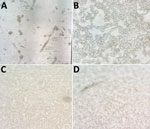
We isolated virus in 13 (81.25%) patients out of 16 infected persons inoculated. From those persons, we recovered MPXV in 17 (80%) of 21 samples assayed: 4 (80%) respiratory swab samples (3 from Vero-E6 and 1 from MRC-5 cells) and 13 (81.25%) lesion swab samples (13 from Vero-E6 cells and 11 from MRC-5 cells) (p = 0.68) (Table 2). In 6 subcultures in A549 cells and in LLC-MK2 cells, cytopathic effect was observed and confirmed by PCR (Figure 1).
We characterized the virus using Sanger sequencing method, and we purified then sequenced the PCR product by using BigDye Terminator v1.1 Cycle Sequencing Kit with an ABI PRISM 3700 DNA analyzer (both ThermoFisher Scientific, https://www.thermofisher.com). We analyzed the sequences subsequently obtained by using IQ-TREE multicore version 2.1.3 (http://www.iqtree.org). We performed tree reconstruction using best-fit model chosen according to Bayesian information criterion. We tested tree branches by the SH-like aLRT method with 1,000 replicates, generating 1,000 samples for ultrafast bootstrap (Figure 2).
In MPXV infection, human-to-human transmission can result from close contact with respiratory secretions, skin lesions of an infected person, or recently contaminated objects (7). In this study, all infected patients were men who had skin lesions. In <50% of cases, patients were experiencing fever, adenopathy, or malaise. Fifteen patients stated they had sex with men or engaged in high-risk sexual practices. In this study, 12 HIV-positive patients were infected with MPXV but did not have higher viral load (4). Only 2 patients were hospitalized. One patient had fever and severe lymphocytosis. The other was a renal transplant patient hospitalized mainly because of his immunosuppressed state. In that patient, the virus was detected for >1 week. In any case, all the patients had a positive outcome.
All viruses were characterized as clade II, similar to findings from other cases in Spain, suggesting the same focus of infection (S. Buenestado et al., unpub. data, https://virological.org/t/updatetwo-draft-genomes-from-madrid-spain-of-the-monkeypoxvirus-2022-outbreak/848). As expected, the disease followed a self-limited course, and no patients experienced severe complications. DNA sequencing also makes it possible to interpret transmission episodes and confirm the existence of endemic variants (8,9). Community transmission data were available for 10 cases. Only 2 were considered secondary cases, indicating that transmission in that environment at that point was not common but had started and could spread.
When possible, blood and pharyngeal or nasopharyngeal swabs were collected, per World Health Organization recommendations (7,10). Blood samples were only collected from 3 patients, and virus was detected in 2 of them at <4 log10 copies/mL. Viremia occurs very early in the course of infection and usually contains a lower viral load than lesions. On the other hand, the normalized viral load was lower in respiratory swab samples than in lesion swab samples, which was to be expected. Patients sought care at a more advanced stage of infection, in which lesions are already present in different phases.
In this study, virus was easily recovered in standard cell culture (VeroE6, MRC-5) from samples with a real-time qPCR cycle threshold of <31 and ≈3.3 log10 copies/103 cells, according to a standard curve (6). Cytopathic effect appeared in <5 days. In addition, subcultures were achieved in other cell lines commonly used in the laboratory (A549 or LLC-MK2). Those data indicate that at higher viral loads, the virus is complete and transmissible, as has been demonstrated with other viruses such as SARS-CoV-2 (11). A limitation of this study was the lack of detailed clinical information for many patients.
In summary, MPXV requires rapid diagnosis and a rapid public health response. The designed real-time qRT-PCR and virus characterization proved very useful in diagnosing mpox and surveillance for MPXV and could aid in controlling the spread of infection and managing outbreaks. Furthermore, the use of culture can help confirm transmission.
Dr. Álvarez-Argüelles is a researcher and microbiologist dedicated to the field of virology. Her primary research interests are viruses that cause clinical infections and the use and application of virus-detection methods.
Acknowledgment
This project was partially funded by Grupin IDI/2021/000033.
References
- World Health Organization. Monkeypox: experts give virus variants new names [cited 2022 Sep 30]. https://www.who.int/news/item/12-08-2022-monkeypox–experts-give-virus-variants-new-names
- Adler H, Gould S, Hine P, Snell LB, Wong W, Houlihan CF, et al.; NHS England High Consequence Infectious Diseases (Airborne) Network. Clinical features and management of human monkeypox: a retrospective observational study in the UK. Lancet Infect Dis. 2022;22:1153–62. DOIPubMedGoogle Scholar
- World Health Organization. Monkeypox—United Kingdom of Great Britain and Northern Ireland [cited 2022 May 18]. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON383
- Iñigo Martínez J, Gil Montalbán E, Jiménez Bueno S, Martín Martínez F, Nieto Juliá A, Sánchez Díaz J, et al. Monkeypox outbreak predominantly affecting men who have sex with men, Madrid, Spain, 26 April to 16 June 2022. Euro Surveill. 2022;27:
2200471 . DOIPubMedGoogle Scholar - Fedele CG, Negredo A, Molero F, Sánchez-Seco MP, Tenorio A. Use of internally controlled real-time genome amplification for detection of variola virus and other orthopoxviruses infecting humans. J Clin Microbiol. 2006;44:4464–70. DOIPubMedGoogle Scholar
- Álvarez-Argüelles ME, de Oña-Navarro M, Rojo-Alba S, Torrens-Muns M, Junquera-Llaneza ML, Antonio-Boga J, et al. Quantification of human papilloma virus (HPV) DNA using the Cobas 4800 system in women with and without pathological alterations attributable to the virus. J Virol Methods. 2015;222:95–102. DOIPubMedGoogle Scholar
- World Health Organization. Laboratory testing for the monkeypox virus. Interim guidance [cited 2022 May 23]. https://apps.who.int/iris/rest/bitstreams/1425052/retrieve
- Nakazawa Y, Emerson GL, Carroll DS, Zhao H, Li Y, Reynolds MG, et al. Phylogenetic and ecologic perspectives of a monkeypox outbreak, southern Sudan, 2005. Emerg Infect Dis. 2013;19:237–45. DOIPubMedGoogle Scholar
- Faye O, Pratt CB, Faye M, Fall G, Chitty JA, Diagne MM, et al. Genomic characterisation of human monkeypox virus in Nigeria. Lancet Infect Dis. 2018;18:246. DOIPubMedGoogle Scholar
- Hasso M, Perusini S, Eshaghi A, Tang E, Olsha R, Zhang H, et al. Monkeypox virus detection in different clinical specimen types. Emerg Infect Dis. 2022;28:2513–5. DOIPubMedGoogle Scholar
- La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–61. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: June 12, 2023
Table of Contents – Volume 29, Number 7—July 2023
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Santiago Melón García, Hospital Universitario Central de Asturias, Microbiology Av. Roma, s/n., Oviedo, Asturias, ES 33011
Top