Volume 29, Number 9—September 2023
Dispatch
Population Analysis of Escherichia coli Sequence Type 361 and Reduced Cefiderocol Susceptibility, France
Figure 2
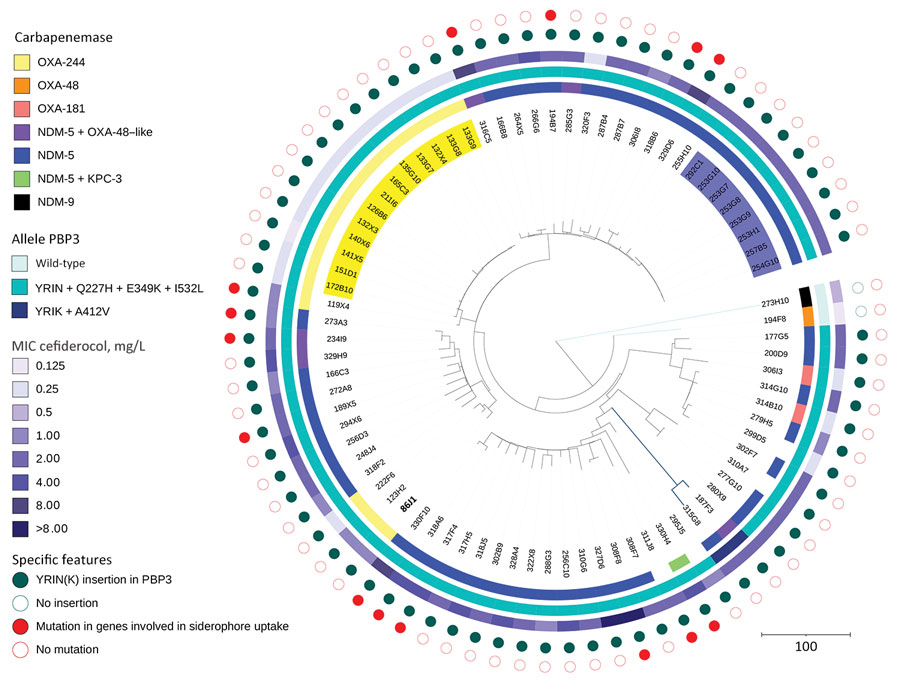
Figure 2. Phylogenetic analysis of 80 Escherichia coli ST361 isolates collected during 2015–2022 and used in a population analysis of E. coli ST361 and reduced cefiderocol susceptibility, France. Isolates were sent to the French National Reference Center for carbapenem-resistant Enterobacterales testing as part of routine surveillance. The phylogenetic tree was built by using SNIppy version 4.6.0 (https://github.com/tseemann/snippy) on whole-genome sequences. Data were visualized using iTOL 6.5.2 (https://itol.embl.de). The most ancient isolate, isolate no. 86J1 collected in 2015 (bold text, lower left of tree), was used as reference genome. A total of 4,957,882 nt positions were analyzed in the comparison. Colors indicate various outbreaks involving 13 OXA-244 producers (yellow) and 7 NDM-5 producers (blue). Two specific features are represented with filled circles: a YRIN(K) insertion in PBP3 (green) and chromosomal mutations within genes involved in siderophore-iron uptake (red). Genes investigated were cirA, fiu, fepA, fepB, fecA, fhuA, tonB, pcnB, exbB, exbD, baeS/baeR, and ompR/envZ. Scale bar indicates nucleotide substitutions per site. KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA, oxacillinase; PBP3, penicillin-binding protein 3.