Volume 30, Number 1—January 2024
Dispatch
Emergence of Novel Norovirus GII.4 Variant
Figure 2
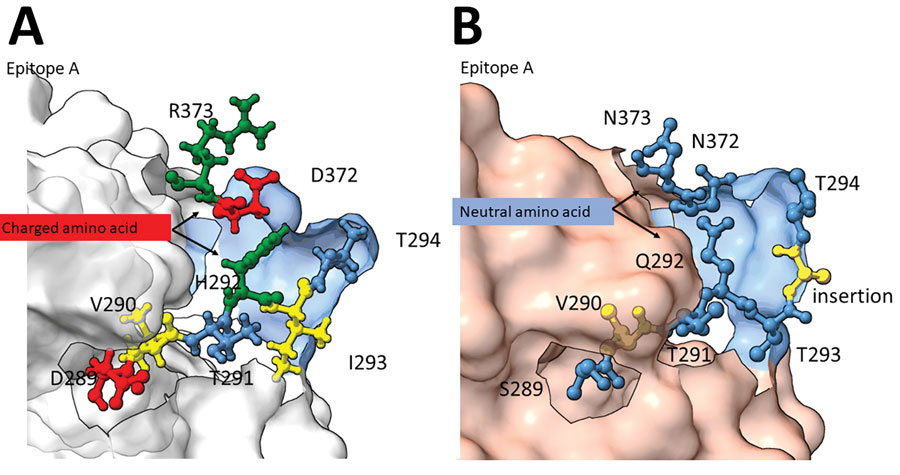
Figure 2. Structural changes of emergent novel norovirus GII.4 strains from 3 continents. A) Sydney GII.4 strain (GenBank accession no. JX459908); B) GII.4 San Francisco strain. The 3-dimensional structure models were predicted by using ChimeraX version 1.4 (11) and the alphafold prediction tool (12). Models show structural changes near and within the epitope A antigenic region on GII.4 San Francisco P-domain (panel B) are overlayed on a GII.4 Sydney 2012 backbone (Protein Data Bank, https://www.rcsb.org/structure/4OP7). Negatively (red) and positively (green) charged amino acids of GII.4 Sydney (panel A) were replaced with neutral amino acids (blue) in the GII.4 San Francisco strain and a hydrophobic (yellow) amino acid, alanine, was inserted between T293 and T294.
References
- Chhabra P, de Graaf M, Parra GI, Chan MC, Green K, Martella V, et al. Updated classification of norovirus genogroups and genotypes. J Gen Virol. 2019;100:1393–406. DOIPubMedGoogle Scholar
- Farahmand M, Moghoofei M, Dorost A, Shoja Z, Ghorbani S, Kiani SJ, et al. Global prevalence and genotype distribution of norovirus infection in children with gastroenteritis: A meta-analysis on 6 years of research from 2015 to 2020. Rev Med Virol. 2022;32:
e2237 . DOIPubMedGoogle Scholar - Parra GI, Tohma K, Ford-Siltz LA, Eguino P, Kendra JA, Pilewski KA, et al. Minimal antigenic evolution after a decade of norovirus GII.4 Sydney_2012 circulation in humans. J Virol. 2023;97:
e0171622 . DOIPubMedGoogle Scholar - Lindesmith LC, Boshier FAT, Brewer-Jensen PD, Roy S, Costantini V, Mallory ML, et al. Immune imprinting drives human norovirus potential for global spread. MBio. 2022;13:
e0186122 . DOIPubMedGoogle Scholar - Cannon JL, Barclay L, Collins NR, Wikswo ME, Castro CJ, Magaña LC, et al. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of novel GII.4 recombinant viruses. J Clin Microbiol. 2017;55:2208–21. DOIPubMedGoogle Scholar
- Manouana GP, Nguema-Moure PA, Mbong Ngwese M, Bock CT, Kremsner PG, Borrmann S, et al. Genetic diversity of enteric viruses in children under five years old in Gabon. Viruses. 2021;13:545. DOIPubMedGoogle Scholar
- Brown JR, Roy S, Ruis C, Yara Romero E, Shah D, Williams R, et al. Norovirus whole-genome sequencing by SureSelect target enrichment: a robust and sensitive method. J Clin Microbiol. 2016;54:2530–7. DOIPubMedGoogle Scholar
- Mans J, Murray TY, Taylor MB. Novel norovirus recombinants detected in South Africa. Virol J. 2014;11:168. DOIPubMedGoogle Scholar
- Parra GI, Squires RB, Karangwa CK, Johnson JA, Lepore CJ, Sosnovtsev SV, et al. Static and evolving norovirus genotypes: implications for epidemiology and immunity. PLoS Pathog. 2017;13:
e1006136 . DOIPubMedGoogle Scholar - Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. DOIPubMedGoogle Scholar
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. DOIPubMedGoogle Scholar
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. DOIPubMedGoogle Scholar
- Widdowson MA, Cramer EH, Hadley L, Bresee JS, Beard RS, Bulens SN, et al. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus—United States, 2002. J Infect Dis. 2004;190:27–36. DOIPubMedGoogle Scholar
- Ruis C, Lindesmith LC, Mallory ML, Brewer-Jensen PD, Bryant JM, Costantini V, et al. Preadaptation of pandemic GII.4 noroviruses in unsampled virus reservoirs years before emergence. Virus Evol. 2020;6:veaa067.
- Debbink K, Donaldson EF, Lindesmith LC, Baric RS. Genetic mapping of a highly variable norovirus GII.4 blockade epitope: potential role in escape from human herd immunity. J Virol. 2012;86:1214–26. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.