Volume 30, Number 10—October 2024
Dispatch
Circovirus Hepatitis in Immunocompromised Patient, Switzerland
Figure 2
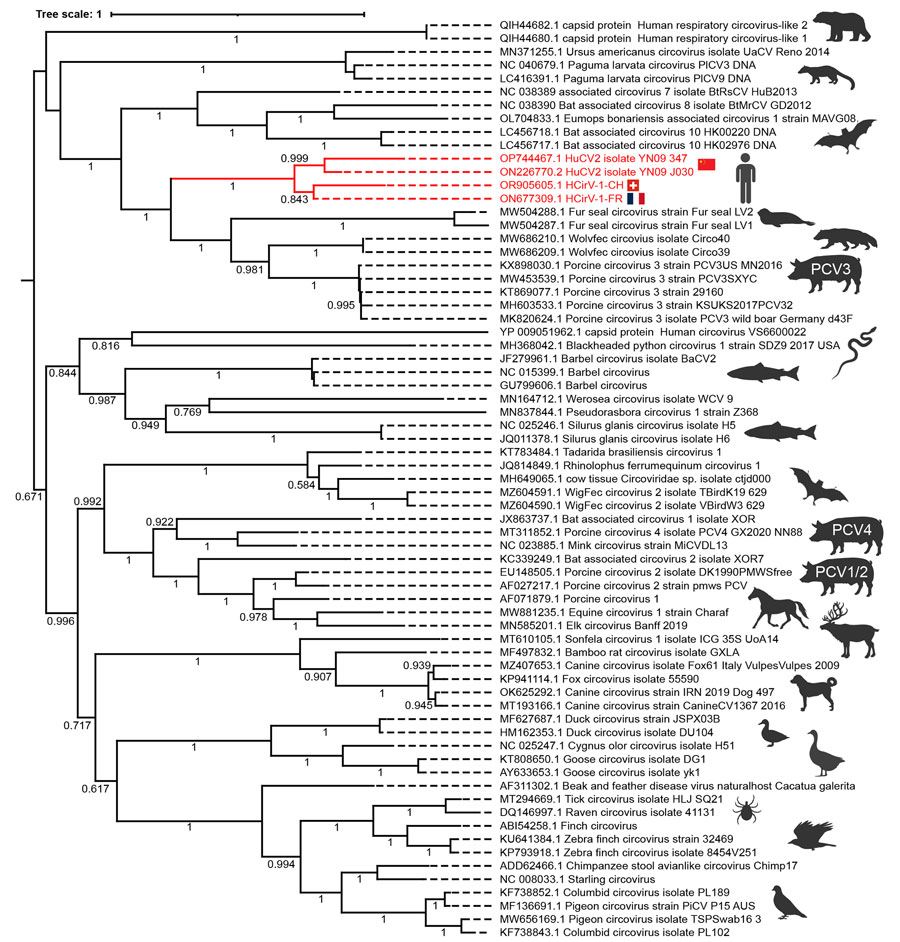
Figure 2. Phylogenetic analysis of the circovirus isolate from a patient with hepatitis of unknown origin, Switzerland (HCirV-1-CH). Capsid protein sequences representative of human and animal circovirus strains are shown. We aligned sequences by using MAFFT (https://mafft.cbrc.jp/alignment/software) under the L-INS-I parameter and performed maximum-likelihood phylogenetic reconstruction through the IQ-Tree portal (http://www.iqtree.org). Red indicates HCirV-1-FR, HCirV-1-CH, and HuCV2 sequences. Tree scale indicates the number of amino acid substitutions per site. Display range for bootstraps is 0.5–1.0. GenBank accession numbers are provided. HCirV-1-CH, human circovirus 1 from patient in Switzerland; HCirV-1-FR, published human circovirus 1 genome from France; HuCV2, human circovirus sequence detected in injection drug users in China (10); PCV, porcine circovirus.
References
- Opriessnig T, Xiao CT, Mueller NJ, Denner J. Emergence of novel circoviruses in humans and pigs and their possible importance for xenotransplantation and blood transfusions. Xenotransplantation. 2024;31:
e12842 . DOIPubMedGoogle Scholar - Kroeger M, Temeeyasen G, Piñeyro PE. Five years of porcine circovirus 3: What have we learned about the clinical disease, immune pathogenesis, and diagnosis. Virus Res. 2022;314:
198764 . DOIPubMedGoogle Scholar - Niu G, Chen S, Li X, Zhang L, Ren L. Advances in crosstalk between porcine circoviruses and host. Viruses. 2022;14:1419. DOIPubMedGoogle Scholar
- Rosell C, Segalés J, Domingo M. Hepatitis and staging of hepatic damage in pigs naturally infected with porcine circovirus type 2. Vet Pathol. 2000;37:687–92. DOIPubMedGoogle Scholar
- Pérot P, Fourgeaud J, Rouzaud C, Regnault B, Da Rocha N, Fontaine H, et al. Circovirus hepatitis infection in heart-lung transplant patient, France. Emerg Infect Dis. 2023;29:286–93. DOIPubMedGoogle Scholar
- Nienhold R, Mensah N, Frank A, Graber A, Koike J, Schwab N, et al. Unbiased screen for pathogens in human paraffin-embedded tissue samples by whole genome sequencing and metagenomics. Front Cell Infect Microbiol. 2022;12:
968135 . DOIPubMedGoogle Scholar - Fourgeaud J, Regnault B, Ok V, Da Rocha N, Sitterlé É, Mekouar M, et al. Performance of clinical metagenomics in France: a prospective observational study. Lancet Microbe. 2024;5:e52–61. DOIPubMedGoogle Scholar
- Pérot P, Bigot T, Temmam S, Regnault B, Eloit M. Microseek: a protein-based metagenomic pipeline for virus diagnostic and discovery. Viruses. 2022;14:1990. DOIPubMedGoogle Scholar
- Li Y, Zhang P, Ye M, Tian RR, Li N, Cao L, et al. Novel circovirus in blood from intravenous drug users, Yunnan, China. Emerg Infect Dis. 2023;29:1015–9. DOIPubMedGoogle Scholar
- Pérot P, Da Rocha N, Farcet MR, Kreil TR, Eloit M. Human Circovirus is not detected in plasma pools for fractionation. Transfusion. 2024;64:16–8. DOIPubMedGoogle Scholar
- Turlewicz-Podbielska H, Augustyniak A, Pomorska-Mól M. Novel porcine circoviruses in view of lessons learned from porcine circovirus type 2-epidemiology and threat to pigs and other species. Viruses. 2022;14:261. DOIPubMedGoogle Scholar
1These first authors contributed equally to this article.
2These senior authors contributed equally to this article.